New species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus shows an ortholog of the E. canis major immunogenic glycoprotein gp36 with a new sequence of tandem repeats
- PMID: 23231731
- PMCID: PMC3533933
- DOI: 10.1186/1756-3305-5-291
New species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus shows an ortholog of the E. canis major immunogenic glycoprotein gp36 with a new sequence of tandem repeats
Abstract
Background: Ehrlichia species are the etiological agents of emerging and life-threatening tick-borne human zoonoses that inflict serious and fatal infections in companion animals and livestock. The aim of this paper was to phylogeneticaly characterise a new species of Ehrlichia isolated from Rhipicephalus (Boophilus) microplus from Minas Gerais, Brazil.
Methods: The agent was isolated from the hemolymph of Rhipicephalus (B.) microplus engorged females that had been collected from naturally infested cattle in a farm in the state of Minas Gerais, Brazil. This agent was then established and cultured in IDE8 tick cells. The molecular and phylogenetic analysis was based on 16S rRNA, groEL, dsb, gltA and gp36 genes. We used the maximum likelihood method to construct the phylogenetic trees.
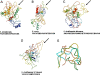
Results: The phylogenetic trees based on 16S rRNA, groEL, dsb and gltA showed that the Ehrlichia spp isolated in this study falls in a clade separated from any previously reported Ehrlichia spp. The molecular analysis of the ortholog of gp36, the major immunoreactive glycoproteins in E. canis and ortholog of the E. chaffeensis gp47, showed a unique tandem repeat of 9 amino acids (VPAASGDAQ) when compared with those reported for E. canis, E. chaffeensis and the related mucin-like protein in E. ruminantium.
Conclusions: Based on the molecular and phylogenetic analysis of the 16S rRNA, groEL, dsb and gltA genes we concluded that this tick-derived microorganism isolated in Brazil is a new species, named E. mineirensis (UFMG-EV), with predicted novel antigenic properties in the gp36 ortholog glycoprotein. Further studies on this new Ehrlichia spp should address questions about its transmissibility by ticks and its pathogenicity for mammalian hosts.
Figures



References
-
- Doyle KC, Labruna MB, Breitschwerdt EB, Tang YW, Corstvet RE, Hegarty BC, Bloch KC, Li P, Walker DH, McBride JW. Detection of medically important Ehrlichia by quantitative multicolor TaqMan real-time polymerase chain reaction of the dsb gene. J Mol Diagn. 2005;7(4):504–510. doi: 10.1016/S1525-1578(10)60581-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

