Dissecting the mechanisms of Notch induced hyperplasia
- PMID: 23232763
- PMCID: PMC3545308
- DOI: 10.1038/emboj.2012.326
Dissecting the mechanisms of Notch induced hyperplasia
Abstract
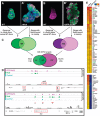
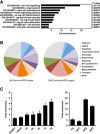
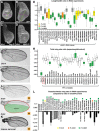
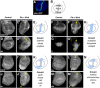
The outcome of the Notch pathway on proliferation depends on cellular context, being growth promotion in some, including several cancers, and growth inhibition in others. Such disparate outcomes are evident in Drosophila wing discs, where Notch overactivation causes hyperplasia despite having localized inhibitory effects on proliferation. To understand the underlying mechanisms, we have used genomic strategies to identify the Notch-CSL target genes directly activated during wing disc hyperplasia. Among them were genes involved in both autonomous and non-autonomous regulation of proliferation, growth and cell death, providing molecular explanations for many characteristics of Notch induced wing disc hyperplasia previously reported. The Notch targets exhibit different response patterns, which are shaped by both positive and negative feed-forward regulation between the Notch targets themselves. We propose, therefore, that both the characteristics of the direct Notch targets and their cross-regulatory relationships are important in coordinating the pattern of hyperplasia.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
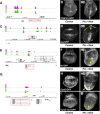
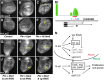
References
-
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg G-H (2007) GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns 7: 323–331 - PubMed
-
- Bachmann A, Knust E (1998) Dissection of cis-regulatory elements of the Drosophila gene Serrate. Dev Genes Evol 208: 346–351 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

