TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells
- PMID: 23233547
- PMCID: PMC3589534
- DOI: 10.1101/gr.147678.112
TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells
Abstract
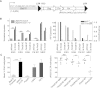
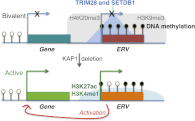
TRIM28 is critical for the silencing of endogenous retroviruses (ERVs) in embryonic stem (ES) cells. Here, we reveal that an essential impact of this process is the protection of cellular gene expression in early embryos from perturbation by cis-acting activators contained within these retroelements. In TRIM28-depleted ES cells, repressive chromatin marks at ERVs are replaced by histone modifications typical of active enhancers, stimulating transcription of nearby cellular genes, notably those harboring bivalent promoters. Correspondingly, ERV-derived sequences can repress or enhance expression from an adjacent promoter in transgenic embryos depending on their TRIM28 sensitivity in ES cells. TRIM28-mediated control of ERVs is therefore crucial not just to prevent retrotransposition, but more broadly to safeguard the transcriptional dynamics of early embryos.
Figures




References
-
- Barde I, Salmon P, Trono D 2010. Production and titration of lentiviral vectors. Curr Protoc Neurosci 53: 4.21.1–4.21.23 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
-
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, de Sio FS, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. 2012. Liver-specific ablation of KRAB associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology 56: 1279–1290 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
