Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression
- PMID: 23233736
- PMCID: PMC3563734
- DOI: 10.1158/0008-5472.CAN-12-2968
Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression
Abstract
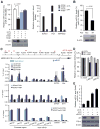
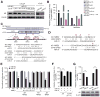
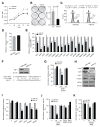
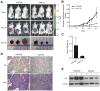
Androgen receptor signaling plays a critical role in prostate cancer pathogenesis. Yet, the regulation of androgen receptor signaling remains elusive. Even with stringent androgen deprivation therapy, androgen receptor signaling persists. Here, our data suggest that there is a complex interaction between the expression of the tumor suppressor miRNA, miR-31, and androgen receptor signaling. We examined primary and metastatic prostate cancer and found that miR-31 expression was reduced as a result of promoter hypermethylation, and importantly, the levels of miR-31 expression were inversely correlated with the aggressiveness of the disease. As the expression of androgen receptor and miR-31 was inversely correlated in the cell lines, our study further suggested that miR-31 and androgen receptor could mutually repress each other. Upregulation of miR-31 effectively suppressed androgen receptor expression through multiple mechanisms and inhibited prostate cancer growth in vivo. Notably, we found that miR-31 targeted androgen receptor directly at a site located in the coding region, which was commonly mutated in prostate cancer. In addition, miR-31 suppressed cell-cycle regulators including E2F1, E2F2, EXO1, FOXM1, and MCM2. Together, our findings suggest a novel androgen receptor regulatory mechanism mediated through miR-31 expression. The downregulation of miR-31 may disrupt cellular homeostasis and contribute to the evolution and progression of prostate cancer. We provide implications for epigenetic treatment and support clinical development of detecting miR-31 promoter methylation as a novel biomarker.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures


References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
-
- Messing EM, Manola J, Sarosdy M, Wilding G, Crawford ED, Trump D. Immediate hormonal therapy compared with observation after radical prostatectomy and pelvic lymphadenectomy in men with node-positive prostate cancer. N Engl J Med. 1999;341:1781–8. - PubMed
-
- Huggins C, Stevens RE, Hodges CV. The effect of castration on advanced carcinoma of the prostate gland. Arch Surg. 1941;43:209–15.
-
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

