On the binding affinity of macromolecular interactions: daring to ask why proteins interact
- PMID: 23235262
- PMCID: PMC3565702
- DOI: 10.1098/rsif.2012.0835
On the binding affinity of macromolecular interactions: daring to ask why proteins interact
Abstract
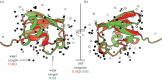
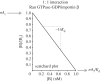
Interactions between proteins are orchestrated in a precise and time-dependent manner, underlying cellular function. The binding affinity, defined as the strength of these interactions, is translated into physico-chemical terms in the dissociation constant (K(d)), the latter being an experimental measure that determines whether an interaction will be formed in solution or not. Predicting binding affinity from structural models has been a matter of active research for more than 40 years because of its fundamental role in drug development. However, all available approaches are incapable of predicting the binding affinity of protein-protein complexes from coordinates alone. Here, we examine both theoretical and experimental limitations that complicate the derivation of structure-affinity relationships. Most work so far has concentrated on binary interactions. Systems of increased complexity are far from being understood. The main physico-chemical measure that relates to binding affinity is the buried surface area, but it does not hold for flexible complexes. For the latter, there must be a significant entropic contribution that will have to be approximated in the future. We foresee that any theoretical modelling of these interactions will have to follow an integrative approach considering the biology, chemistry and physics that underlie protein-protein recognition.
Figures







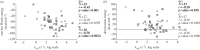
References
-
- Mulder GJ. 1839. Ueber die Zusammensetzung einiger thierischen Substanzen. J. Prakt. Chem. 16, 129–15210.1002/prac.18390160137 (doi:10.1002/prac.18390160137) - DOI - DOI
-
- Perrett D. 2007. From ‘protein’ to the beginnings of clinical proteomics. Proteomics Clin. Appl. 1, 720–73810.1002/prca.200700525 (doi:10.1002/prca.200700525) - DOI - DOI - PubMed
-
- Tanford C, Reynolds J. 2004. Nature‘s robots: a history of proteins. New York, NY: Oxford University Press
-
- Proust LJ. 1819. Recherches sur le principe qui assaisonne les fromages. Ann. Chim. Phys. 29, 29–49
-
- Meyer CE, Rose WC. 1936. The spatial configuration of a-amino-b-hydroxy-n-butyric acid. J. Biol. Chem. 115, 721–729
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
