Estimating the rate of intersubtype recombination in early HIV-1 group M strains
- PMID: 23236072
- PMCID: PMC3571495
- DOI: 10.1128/JVI.02478-12
Estimating the rate of intersubtype recombination in early HIV-1 group M strains
Abstract
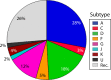
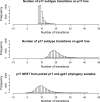
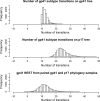
West Central Africa has been implicated as the epicenter of the HIV-1 epidemic, and almost all group M subtypes can be found there. Previous analysis of early HIV-1 group M sequences from Kinshasa in the Democratic Republic of Congo, formerly Zaire, revealed that isolates from a number of individuals fall in different positions in phylogenetic trees constructed from sequences from opposite ends of the genome as a result of recombination between viruses of different subtypes. Here, we use discrete ancestral trait mapping to develop a procedure for quantifying HIV-1 group M intersubtype recombination across phylogenies, using individuals' gag (p17) and env (gp41) subtypes. The method was applied to previously described HIV-1 group M sequences from samples obtained in Kinshasa early in the global radiation of HIV. Nine different p17 and gp41 intersubtype recombinant combinations were present in the data set. The mean number of excess ancestral subtype transitions (NEST) required to map individuals' p17 subtypes onto the gp14 phylogeny samples, compared to the number required to map them onto the p17 phylogenies, and vice versa, indicated that excess subtype transitions occurred at a rate of approximately 7 × 10(-3) to 8 × 10(-3) per lineage per year as a result of intersubtype recombination. Our results imply that intersubtype recombination may have occurred in approximately 20% of lineages evolving over a period of 30 years and confirm intersubtype recombination as a substantial force in generating HIV-1 group M diversity.
Figures

References
-
- Gao F, Bailes E, Robertson DL, Chen YL, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 - PubMed
-
- Plantier J-C, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871–872 - PubMed
-
- Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. 2000. HIV-1 nomenclature proposal. Science 288:55–57 - PubMed
-
- Rambaut A, Posada D, Crandall KA, Holmes EC. 2004. The causes and consequences of HIV evolution. Nat. Rev. Genet. 5:52–61 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

