Cross-biome metagenomic analyses of soil microbial communities and their functional attributes
- PMID: 23236140
- PMCID: PMC3535587
- DOI: 10.1073/pnas.1215210110
Cross-biome metagenomic analyses of soil microbial communities and their functional attributes
Abstract
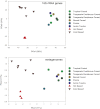
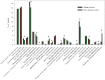
For centuries ecologists have studied how the diversity and functional traits of plant and animal communities vary across biomes. In contrast, we have only just begun exploring similar questions for soil microbial communities despite soil microbes being the dominant engines of biogeochemical cycles and a major pool of living biomass in terrestrial ecosystems. We used metagenomic sequencing to compare the composition and functional attributes of 16 soil microbial communities collected from cold deserts, hot deserts, forests, grasslands, and tundra. Those communities found in plant-free cold desert soils typically had the lowest levels of functional diversity (diversity of protein-coding gene categories) and the lowest levels of phylogenetic and taxonomic diversity. Across all soils, functional beta diversity was strongly correlated with taxonomic and phylogenetic beta diversity; the desert microbial communities were clearly distinct from the nondesert communities regardless of the metric used. The desert communities had higher relative abundances of genes associated with osmoregulation and dormancy, but lower relative abundances of genes associated with nutrient cycling and the catabolism of plant-derived organic compounds. Antibiotic resistance genes were consistently threefold less abundant in the desert soils than in the nondesert soils, suggesting that abiotic conditions, not competitive interactions, are more important in shaping the desert microbial communities. As the most comprehensive survey of soil taxonomic, phylogenetic, and functional diversity to date, this study demonstrates that metagenomic approaches can be used to build a predictive understanding of how microbial diversity and function vary across terrestrial biomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Fierer N, Ladau J. Predicting microbial distributions in space and time. Nat Methods. 2012;9(6):549–551. - PubMed
-
- Rousk J, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4(10):1340–1351. - PubMed
-
- Osborne CA, Zwart AB, Broadhurst LM, Young AG, Richardson AE. The influence of sampling strategies and spatial variation on the detected soil bacterial communities under three different land-use types. FEMS Microbiol Ecol. 2011;78(1):70–79. - PubMed
-
- Chu H, et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12(11):2998–3006. - PubMed
-
- Kuramae EE, et al. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol Ecol. 2012;79(1):12–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

