Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes
- PMID: 23236188
- PMCID: PMC3535588
- DOI: 10.1073/pnas.1213818110
Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes
Abstract
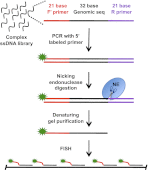
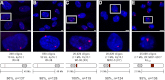
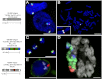
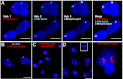
A host of observations demonstrating the relationship between nuclear architecture and processes such as gene expression have led to a number of new technologies for interrogating chromosome positioning. Whereas some of these technologies reconstruct intermolecular interactions, others have enhanced our ability to visualize chromosomes in situ. Here, we describe an oligonucleotide- and PCR-based strategy for fluorescence in situ hybridization (FISH) and a bioinformatic platform that enables this technology to be extended to any organism whose genome has been sequenced. The oligonucleotide probes are renewable, highly efficient, and able to robustly label chromosomes in cell culture, fixed tissues, and metaphase spreads. Our method gives researchers precise control over the sequences they target and allows for single and multicolor imaging of regions ranging from tens of kilobases to megabases with the same basic protocol. We anticipate this technology will lead to an enhanced ability to visualize interphase and metaphase chromosomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

