Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi
- PMID: 23236275
- PMCID: PMC3516569
- DOI: 10.1371/journal.ppat.1003037
Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi
Abstract
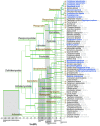
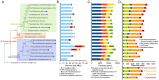
The class Dothideomycetes is one of the largest groups of fungi with a high level of ecological diversity including many plant pathogens infecting a broad range of hosts. Here, we compare genome features of 18 members of this class, including 6 necrotrophs, 9 (hemi)biotrophs and 3 saprotrophs, to analyze genome structure, evolution, and the diverse strategies of pathogenesis. The Dothideomycetes most likely evolved from a common ancestor more than 280 million years ago. The 18 genome sequences differ dramatically in size due to variation in repetitive content, but show much less variation in number of (core) genes. Gene order appears to have been rearranged mostly within chromosomal boundaries by multiple inversions, in extant genomes frequently demarcated by adjacent simple repeats. Several Dothideomycetes contain one or more gene-poor, transposable element (TE)-rich putatively dispensable chromosomes of unknown function. The 18 Dothideomycetes offer an extensive catalogue of genes involved in cellulose degradation, proteolysis, secondary metabolism, and cysteine-rich small secreted proteins. Ancestors of the two major orders of plant pathogens in the Dothideomycetes, the Capnodiales and Pleosporales, may have had different modes of pathogenesis, with the former having fewer of these genes than the latter. Many of these genes are enriched in proximity to transposable elements, suggesting faster evolution because of the effects of repeat induced point (RIP) mutations. A syntenic block of genes, including oxidoreductases, is conserved in most Dothideomycetes and upregulated during infection in L. maculans, suggesting a possible function in response to oxidative stress.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans-Leptosphaeria biglobosa species complex of fungal pathogens.BMC Genomics. 2014 Oct 12;15:891. doi: 10.1186/1471-2164-15-891. BMC Genomics. 2014. PMID: 25306241 Free PMC article.
-
Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis.PLoS Genet. 2011 Jun;7(6):e1002070. doi: 10.1371/journal.pgen.1002070. Epub 2011 Jun 9. PLoS Genet. 2011. PMID: 21695235 Free PMC article.
-
The landscape of transposable elements in the finished genome of the fungal wheat pathogen Mycosphaerella graminicola.BMC Genomics. 2014 Dec 17;15(1):1132. doi: 10.1186/1471-2164-15-1132. BMC Genomics. 2014. PMID: 25519841 Free PMC article.
-
Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi.FEMS Microbiol Rev. 2013 Jan;37(1):67-93. doi: 10.1111/j.1574-6976.2012.00349.x. Epub 2012 Aug 29. FEMS Microbiol Rev. 2013. PMID: 22931103 Review.
-
Karyotype Variability in Plant-Pathogenic Fungi.Annu Rev Phytopathol. 2017 Aug 4;55:483-503. doi: 10.1146/annurev-phyto-080615-095928. Annu Rev Phytopathol. 2017. PMID: 28777924 Review.
Cited by
-
Relative abundance of and composition within fungal orders differ between cheatgrass (Bromus tectorum) and sagebrush (Artemisia tridentate)-associated soils.PLoS One. 2015 Jan 28;10(1):e0117026. doi: 10.1371/journal.pone.0117026. eCollection 2015. PLoS One. 2015. PMID: 25629158 Free PMC article.
-
The northern corn leaf blight resistance gene Ht1 encodes an nucleotide-binding, leucine-rich repeat immune receptor.Mol Plant Pathol. 2023 Jul;24(7):758-767. doi: 10.1111/mpp.13267. Epub 2022 Sep 30. Mol Plant Pathol. 2023. PMID: 36180934 Free PMC article.
-
Genome and transcriptome reveal lithophilic adaptation of Cladophialophora brunneola, a new rock-inhabiting fungus.Mycology. 2024 Jan 2;14(4):326-343. doi: 10.1080/21501203.2023.2256764. eCollection 2023. Mycology. 2024. PMID: 38187882 Free PMC article.
-
Genomic biosurveillance of forest invasive alien enemies: A story written in code.Evol Appl. 2019 Sep 10;13(1):95-115. doi: 10.1111/eva.12853. eCollection 2020 Jan. Evol Appl. 2019. PMID: 31892946 Free PMC article. Review.
-
Wheat Straw Return Influences Nitrogen-Cycling and Pathogen Associated Soil Microbiota in a Wheat-Soybean Rotation System.Front Microbiol. 2019 Aug 8;10:1811. doi: 10.3389/fmicb.2019.01811. eCollection 2019. Front Microbiol. 2019. PMID: 31440226 Free PMC article.
References
-
- Kirk P, Cannon P, Minter D, Stalpers J (2008) Ainsworth and Bisby's dictionary of the Fungi, 10th ed. Wallingford, UK: CAB International.
-
- Ewaze JO, Summerbell RC, Scott JA (2008) Ethanol physiology in the warehouse-staining fungus, Baudoinia compniacensis. Mycol Res 112: 1373–1380. - PubMed
-
- Gioulekas D, Damialis A, Papakosta D, Spieksma F, Giouleka P, et al. (2004) Allergenic fungi spore records (15 years) and sensitization in patients with respiratory allergy in Thessaloniki-Greece. J Investig Allergol Clin Immunol 14: 225–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

