Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach
- PMID: 23236287
- PMCID: PMC3516541
- DOI: 10.1371/journal.pgen.1003065
Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach
Abstract
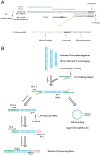
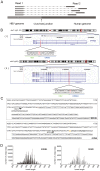
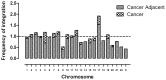
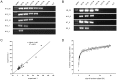
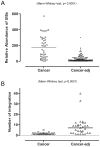
Integration of the viral DNA into host chromosomes was found in most of the hepatitis B virus (HBV)-related hepatocellular carcinomas (HCCs). Here we devised a massive anchored parallel sequencing (MAPS) method using next-generation sequencing to isolate and sequence HBV integrants. Applying MAPS to 40 pairs of HBV-related HCC tissues (cancer and adjacent tissues), we identified 296 HBV integration events corresponding to 286 unique integration sites (UISs) with precise HBV-Human DNA junctions. HBV integration favored chromosome 17 and preferentially integrated into human transcript units. HBV targeted genes were enriched in GO terms: cAMP metabolic processes, T cell differentiation and activation, TGF beta receptor pathway, ncRNA catabolic process, and dsRNA fragmentation and cellular response to dsRNA. The HBV targeted genes include 7 genes (PTPRJ, CNTN6, IL12B, MYOM1, FNDC3B, LRFN2, FN1) containing IPR003961 (Fibronectin, type III domain), 7 genes (NRG3, MASP2, NELL1, LRP1B, ADAM21, NRXN1, FN1) containing IPR013032 (EGF-like region, conserved site), and three genes (PDE7A, PDE4B, PDE11A) containing IPR002073 (3', 5'-cyclic-nucleotide phosphodiesterase). Enriched pathways include hsa04512 (ECM-receptor interaction), hsa04510 (Focal adhesion), and hsa04012 (ErbB signaling pathway). Fewer integration events were found in cancers compared to cancer-adjacent tissues, suggesting a clonal expansion model in HCC development. Finally, we identified 8 genes that were recurrent target genes by HBV integration including fibronectin 1 (FN1) and telomerase reverse transcriptase (TERT1), two known recurrent target genes, and additional novel target genes such as SMAD family member 5 (SMAD5), phosphatase and actin regulator 4 (PHACTR4), and RNA binding protein fox-1 homolog (C. elegans) 1 (RBFOX1). Integrating analysis with recently published whole-genome sequencing analysis, we identified 14 additional recurrent HBV target genes, greatly expanding the HBV recurrent target list. This global survey of HBV integration events, together with recently published whole-genome sequencing analyses, furthered our understanding of the HBV-related HCC.
Conflict of interest statement
Dong Ding is currently employed by Hangzhou Proprium Biotech. This research was support partially by funding from the HPB.
Figures



References
-
- Parkin DM (2006) The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118: 3030–3044. - PubMed
-
- Szmuness W (1978) Hepatocellular carcinoma and the hepatitis B virus: evidence for a causal association. Prog Med Virol 24: 40–69. - PubMed
-
- Feitelson MA, Lee J (2007) Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett 252: 157–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

