GTSE1 is a microtubule plus-end tracking protein that regulates EB1-dependent cell migration
- PMID: 23236459
- PMCID: PMC3517537
- DOI: 10.1371/journal.pone.0051259
GTSE1 is a microtubule plus-end tracking protein that regulates EB1-dependent cell migration
Abstract
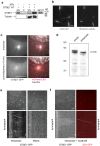
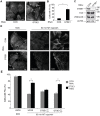
The regulation of cell migration is a highly complex process that is often compromised when cancer cells become metastatic. The microtubule cytoskeleton is necessary for cell migration, but how microtubules and microtubule-associated proteins regulate multiple pathways promoting cell migration remains unclear. Microtubule plus-end binding proteins (+TIPs) are emerging as important players in many cellular functions, including cell migration. Here we identify a +TIP, GTSE1, that promotes cell migration. GTSE1 accumulates at growing microtubule plus ends through interaction with the EB1+TIP. The EB1-dependent +TIP activity of GTSE1 is required for cell migration, as well as for microtubule-dependent disassembly of focal adhesions. GTSE1 protein levels determine the migratory capacity of both nontransformed and breast cancer cell lines. In breast cancers, increased GTSE1 expression correlates with invasive potential, tumor stage, and time to distant metastasis, suggesting that misregulation of GTSE1 expression could be associated with increased invasive potential.
Conflict of interest statement
Figures




References
-
- Zhao X, Guan J-L (2011) Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Advanced Drug Delivery Reviews 63: 610–615 doi:10.1016/j.addr.2010.11.001. - DOI - PMC - PubMed
-
- Li D-M, Feng Y-M (2011) Signaling mechanism of cell adhesion molecules in breast cancer metastasis: potential therapeutic targets. Breast Cancer Res Treat 128: 7–21 doi:10.1007/s10549-011-1499-x. - DOI - PubMed
-
- Thiery JP (2002) Epithelial–mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454 doi:10.1038/nrc822. - DOI - PubMed
-
- Palmer TD, Ashby WJ, Lewis JD, Zijlstra A (2011) Targeting tumor cell motility to prevent metastasis. Advanced Drug Delivery Reviews 63: 568–581 doi:10.1016/j.addr.2011.04.008. - DOI - PMC - PubMed
-
- Kaverina I, Straube A (2011) Regulation of cell migration by dynamic microtubules. Seminars in Cell and Developmental Biology 22: 968–974 doi:10.1016/j.semcdb.2011.09.017. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

