Col1A1 production and apoptotic resistance in TGF-β1-induced epithelial-to-mesenchymal transition-like phenotype of 603B cells
- PMID: 23236489
- PMCID: PMC3517566
- DOI: 10.1371/journal.pone.0051371
Col1A1 production and apoptotic resistance in TGF-β1-induced epithelial-to-mesenchymal transition-like phenotype of 603B cells
Abstract
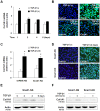
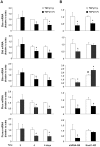
Recent studies have suggested that proliferating cholangiocytes have an important role in the induction of fibrosis, either directly via epithelial-to-mesenchymal transition (EMT), or indirectly via activation of other liver cell types. Transforming growth factor beta 1 (TGF-β1), a critical fibrotic cytokine for hepatic fibrosis, is a potent EMT inducer. This study aimed to clarify the potential contributions of TGF-β1-induced EMT-like cholangiocyte phenotype to collagen production and cell survival of cholangiocytes in vitro. Mouse cholangiocytes (603B cells) were treated with TGF-β1 and EMT-like phenotype alterations were monitored by morphological changes and expression of EMT-associated genes. Alterations in Col1A1 gene, Col1A1-associated miR-29s, and pro-apoptotic genes were measured in TGF-β1-treated 603B cells. Snail1 knockdown was achieved using shRNA to evaluate the contribution of EMT-associated changes to Col1A1 production and cell survival. We found TGF-β1 treatment induced partial EMT-like phenotype transition in 603B cells in a Snail1-dependent manner. TGF-β1 also stimulated collagen α1(I) expression in 603B cells. However, this induction was not parallel to the EMT-like alterations and independent of Snail1 or miR-29 expression. Cells undergoing EMT-like changes showed a modest down-regulation of multiple pro-apoptotic genes and displayed resistance to TNF-α-induced apoptosis. TGF-β1-induced apoptosis resistance was attenuated in Snail1 knockdown 603B cells. TGF-β1-induced Col1A1 production seems to be independent of EMT-like transition and miR-29 expression. Nevertheless, TGF-β1-induced EMT may contribute to the increased survival capacity of cholangiocytes via modulating the expression of pro-apoptotic genes.
Conflict of interest statement
Figures




Similar articles
-
Micro-vesicles from mesenchymal stem cells over-expressing miR-34a inhibit transforming growth factor-β1-induced epithelial-mesenchymal transition in renal tubular epithelial cells in vitro.Chin Med J (Engl). 2020 Apr 5;133(7):800-807. doi: 10.1097/CM9.0000000000000720. Chin Med J (Engl). 2020. PMID: 32149762 Free PMC article.
-
Akt2 mediates TGF-β1-induced epithelial to mesenchymal transition by deactivating GSK3β/snail signaling pathway in renal tubular epithelial cells.Cell Physiol Biochem. 2014;34(2):368-82. doi: 10.1159/000363006. Epub 2014 Jul 11. Cell Physiol Biochem. 2014. PMID: 25059120
-
MiR-30c protects diabetic nephropathy by suppressing epithelial-to-mesenchymal transition in db/db mice.Aging Cell. 2017 Apr;16(2):387-400. doi: 10.1111/acel.12563. Epub 2017 Jan 27. Aging Cell. 2017. PMID: 28127848 Free PMC article.
-
Transforming growth factor-β1 induced epithelial-mesenchymal transition in hepatic fibrosis.Hepatogastroenterology. 2012 Sep;59(118):1960-3. doi: 10.5754/hge11750. Hepatogastroenterology. 2012. PMID: 22369740 Review.
-
Cytokines as drivers: Unraveling the mechanisms of epithelial-mesenchymal transition in COVID-19 lung fibrosis.Biochem Biophys Res Commun. 2023 Dec 17;686:149118. doi: 10.1016/j.bbrc.2023.10.050. Epub 2023 Oct 14. Biochem Biophys Res Commun. 2023. PMID: 37931361 Review.
Cited by
-
A New TGF-β1 Inhibitor, CTI-82, Antagonizes Epithelial-Mesenchymal Transition through Inhibition of Phospho-SMAD2/3 and Phospho-ERK.Biology (Basel). 2020 Jun 28;9(7):143. doi: 10.3390/biology9070143. Biology (Basel). 2020. PMID: 32605257 Free PMC article.
-
Apoptotic biliary epithelial cells and gut dysbiosis in the induction of murine primary biliary cholangitis.J Transl Autoimmun. 2022 Dec 21;6:100182. doi: 10.1016/j.jtauto.2022.100182. eCollection 2023. J Transl Autoimmun. 2022. PMID: 36619656 Free PMC article.
-
Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells.Cancer Cell Int. 2017 Jul 28;17:73. doi: 10.1186/s12935-017-0443-5. eCollection 2017. Cancer Cell Int. 2017. PMID: 28775672 Free PMC article.
-
NOTCH3 Overexpression and Posttranscriptional Regulation by miR-150 Were Associated With EGFR-TKI Resistance in Lung Adenocarcinoma.Oncol Res. 2019 Jul 12;27(7):751-761. doi: 10.3727/096504018X15372657298381. Epub 2019 Feb 7. Oncol Res. 2019. PMID: 30732676 Free PMC article.
-
HK3 overexpression associated with epithelial-mesenchymal transition in colorectal cancer.BMC Genomics. 2018 Feb 9;19(Suppl 3):113. doi: 10.1186/s12864-018-4477-4. BMC Genomics. 2018. PMID: 29504907 Free PMC article.
References
-
- Beaussier M, Wendum D, Schiffer E, Dumont S, Rey C, et al. (2007) Prominent contribution of portal mesenchymal cells to liver fibrosis in ischemic and obstructive cholestatic injuries. Lab Invest 87: 292–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

