X-linked gene transcription patterns in female and male in vivo, in vitro and cloned porcine individual blastocysts
- PMID: 23236494
- PMCID: PMC3517569
- DOI: 10.1371/journal.pone.0051398
X-linked gene transcription patterns in female and male in vivo, in vitro and cloned porcine individual blastocysts
Abstract
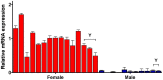
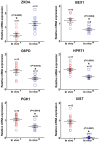
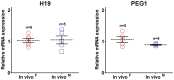
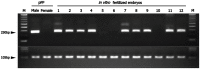
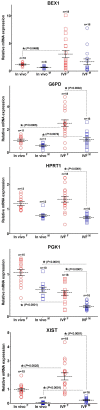
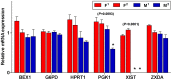
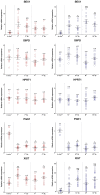
To determine the presence of sexual dimorphic transcription and how in vitro culture environments influence X-linked gene transcription patterns in preimplantation embryos, we analyzed mRNA expression levels in in vivo-derived, in vitro-fertilized (IVF), and cloned porcine blastocysts. Our results clearly show that sex-biased expression occurred between female and male in vivo blastocysts in X-linked genes. The expression levels of XIST, G6PD, HPRT1, PGK1, and BEX1 were significantly higher in female than in male blastocysts, but ZXDA displayed higher levels in male than in female blastocysts. Although we found aberrant expression patterns for several genes in IVF and cloned blastocysts, similar sex-biased expression patterns (on average) were observed between the sexes. The transcript levels of BEX1 and XIST were upregulated and PGK1 was downregulated in both IVF and cloned blastocysts compared with in vivo counterparts. Moreover, a remarkable degree of expression heterogeneity was observed among individual cloned embryos (the level of heterogeneity was similar in both sexes) but only a small proportion of female IVF embryos exhibited variability, indicating that this phenomenon may be primarily caused by faulty reprogramming by the somatic cell nuclear transfer (SCNT) process rather than in vitro conditions. Aberrant expression patterns in cloned embryos of both sexes were not ameliorated by treatment with Scriptaid as a potent HDACi, although the blastocyst rate increased remarkably after this treatment. Taken together, these results indicate that female and male porcine blastocysts produced in vivo and in vitro transcriptional sexual dimorphisms in the selected X-linked genes and compensation of X-linked gene dosage may not occur at the blastocyst stage. Moreover, altered X-linked gene expression frequently occurred in porcine IVF and cloned embryos, indicating that X-linked gene regulation is susceptible to in vitro culture and the SCNT process, which may eventually lead to problems with embryonic or placental defects.
Conflict of interest statement
Figures


Similar articles
-
Oxamflatin treatment enhances cloned porcine embryo development and nuclear reprogramming.Cell Reprogram. 2015 Feb;17(1):28-40. doi: 10.1089/cell.2014.0075. Epub 2014 Dec 30. Cell Reprogram. 2015. PMID: 25548976 Free PMC article.
-
Aggregation of cloned embryos in empty zona pellucida improves derivation efficiency of pig ES-like cells.Zygote. 2016 Dec;24(6):909-917. doi: 10.1017/S0967199416000241. Epub 2016 Oct 3. Zygote. 2016. PMID: 27692031
-
Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos.Cell Reprogram. 2010 Feb;12(1):75-83. doi: 10.1089/cell.2009.0038. Cell Reprogram. 2010. PMID: 20132015 Free PMC article.
-
Scriptaid improves in vitro development and nuclear reprogramming of somatic cell nuclear transfer bovine embryos.Cell Reprogram. 2011 Oct;13(5):431-9. doi: 10.1089/cell.2011.0024. Epub 2011 Jul 20. Cell Reprogram. 2011. PMID: 21774687
-
Scriptaid Treatment Decreases DNA Methyltransferase 1 Expression by Induction of MicroRNA-152 Expression in Porcine Somatic Cell Nuclear Transfer Embryos.PLoS One. 2015 Aug 11;10(8):e0134567. doi: 10.1371/journal.pone.0134567. eCollection 2015. PLoS One. 2015. PMID: 26261994 Free PMC article.
Cited by
-
Effects of RNAi-mediated knockdown of Xist on the developmental efficiency of cloned male porcine embryos.J Reprod Dev. 2016 Dec 20;62(6):591-597. doi: 10.1262/jrd.2016-095. Epub 2016 Aug 29. J Reprod Dev. 2016. PMID: 27569767 Free PMC article.
-
Aberrant expression of Xist in aborted porcine fetuses derived from somatic cell nuclear transfer embryos.Int J Mol Sci. 2014 Nov 25;15(12):21631-43. doi: 10.3390/ijms151221631. Int J Mol Sci. 2014. PMID: 25429426 Free PMC article.
-
Pluripotency and X chromosome dynamics revealed in pig pre-gastrulating embryos by single cell analysis.Nat Commun. 2019 Jan 30;10(1):500. doi: 10.1038/s41467-019-08387-8. Nat Commun. 2019. PMID: 30700715 Free PMC article.
-
Enhancement of Chromatin and Epigenetic Reprogramming in Porcine SCNT Embryos-Progresses and Perspectives.Front Cell Dev Biol. 2022 Jul 11;10:940197. doi: 10.3389/fcell.2022.940197. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35898400 Free PMC article.
-
Amino acid supplementation affects imprinted gene transcription patterns in parthenogenetic porcine blastocysts.PLoS One. 2014 Sep 2;9(9):e106549. doi: 10.1371/journal.pone.0106549. eCollection 2014. PLoS One. 2014. PMID: 25180972 Free PMC article.
References
-
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N (1996) Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. - PubMed
-
- Latham KE (2005) X chromosome imprinting and inactivation in preimplantation mammalian embryos. Trends Genet 21: 120–127. - PubMed
-
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, et al. (2011) Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472: 370–374. - PubMed
-
- Kobayashi S, Isotani A, Mise N, Yamamoto M, Fujihara Y, et al. (2006) Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr Biol 16: 166–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

