Whole-genome reconstruction and mutational signatures in gastric cancer
- PMID: 23237666
- PMCID: PMC4056366
- DOI: 10.1186/gb-2012-13-12-r115
Whole-genome reconstruction and mutational signatures in gastric cancer
Abstract
Background: Gastric cancer is the second highest cause of global cancer mortality. To explore the complete repertoire of somatic alterations in gastric cancer, we combined massively parallel short read and DNA paired-end tag sequencing to present the first whole-genome analysis of two gastric adenocarcinomas, one with chromosomal instability and the other with microsatellite instability.
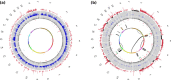
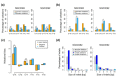
Results: Integrative analysis and de novo assemblies revealed the architecture of a wild-type KRAS amplification, a common driver event in gastric cancer. We discovered three distinct mutational signatures in gastric cancer--against a genome-wide backdrop of oxidative and microsatellite instability-related mutational signatures, we identified the first exome-specific mutational signature. Further characterization of the impact of these signatures by combining sequencing data from 40 complete gastric cancer exomes and targeted screening of an additional 94 independent gastric tumors uncovered ACVR2A, RPL22 and LMAN1 as recurrently mutated genes in microsatellite instability-positive gastric cancer and PAPPA as a recurrently mutated gene in TP53 wild-type gastric cancer.
Conclusions: These results highlight how whole-genome cancer sequencing can uncover information relevant to tissue-specific carcinogenesis that would otherwise be missed from exome-sequencing data.
Figures

References
-
- Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;13:302–308. - PubMed
-
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordonez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;13:184–190. doi: 10.1038/nature08629. - DOI - PMC - PubMed
-
- Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, Ha C, Johnson S, Kennemer MI, Mohan S, Nazarenko I, Watanabe C, Sparks AB, Shames DS, Gentleman R, de Sauvage FJ, Stern H, Pandita A, Ballinger DG, Drmanac R, Modrusan Z, Seshagiri S, Zhang Z. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;13:473–477. doi: 10.1038/nature09004. - DOI - PubMed
-
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;13:191–196. doi: 10.1038/nature08658. - DOI - PMC - PubMed
-
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, Lopez C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernandez JM, Puente DA, Freije JM, Velasco G, Gutierrez-Fernandez A, Costa D, Carrio A, Guijarro S, Enjuanes A. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;13:101–105. doi: 10.1038/nature10113. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

