Vimentin DNA methylation predicts survival in breast cancer
- PMID: 23239149
- PMCID: PMC3838916
- DOI: 10.1007/s10549-012-2353-5
Vimentin DNA methylation predicts survival in breast cancer
Abstract
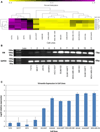
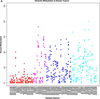
The Vimentin gene plays a pivotal role in epithelial-to-mesenchymal transition and is known to be overexpressed in the prognostically poor basal-like breast cancer subtype. Recent studies have reported Vimentin DNA methylation in association with poor clinical outcomes in other solid tumors, but not in breast cancer. We therefore quantified Vimentin DNA methylation using MALDI-TOF mass spectrometry in breast tumors and matched normal pairs in association with gene expression and survival in a hospital-based study of breast cancer patients. Gene expression data via qRT-PCR in cell lines and oligomicroarray data from breast tissues were correlated with percent methylation in the Vimentin promoter. A threshold of 20 percent average methylation compared with matched normal pairs was set for bivariate and multivariate tests of association between methylation and tumor subtype, tumor histopathology, and survival. Vimentin was differentially methylated in luminal breast cancer cell lines, and in luminal A, luminal B, and HER2-enriched breast tumor subtypes, but was rare in basal-like cell lines and tumors. Increased methylation was strongly correlated with decreased mRNA expression in cell lines, and had a moderate inverse correlation in breast tumors. Vimentin methylation predicted poor overall survival independent of race, subtype, stage, nodal status, or metastatic disease and holds promise as a new prognostic biomarker for breast cancer patients.
Conflict of interest statement
Figures



References
-
- Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, Pretlow TP, Lutterbaugh J, Kasturi L, Willson JK, Rao JS, Shuber A, Markowitz SD. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. - PubMed
-
- Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, Rodrigues A, Magalhaes JS, Oliveira J, Lothe RA, Teixeira MR, Jeronimo C, Lind GE. Three epigenetic biomarkers, GDF15, TMEFF2, and, VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16:5842–5851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

