CS-AMPPred: an updated SVM model for antimicrobial activity prediction in cysteine-stabilized peptides
- PMID: 23240023
- PMCID: PMC3519874
- DOI: 10.1371/journal.pone.0051444
CS-AMPPred: an updated SVM model for antimicrobial activity prediction in cysteine-stabilized peptides
Abstract
The antimicrobial peptides (AMP) have been proposed as an alternative to control resistant pathogens. However, due to multifunctional properties of several AMP classes, until now there has been no way to perform efficient AMP identification, except through in vitro and in vivo tests. Nevertheless, an indication of activity can be provided by prediction methods. In order to contribute to the AMP prediction field, the CS-AMPPred (Cysteine-Stabilized Antimicrobial Peptides Predictor) is presented here, consisting of an updated version of the Support Vector Machine (SVM) model for antimicrobial activity prediction in cysteine-stabilized peptides. The CS-AMPPred is based on five sequence descriptors: indexes of (i) α-helix and (ii) loop formation; and averages of (iii) net charge, (iv) hydrophobicity and (v) flexibility. CS-AMPPred was based on 310 cysteine-stabilized AMPs and 310 sequences extracted from PDB. The polynomial kernel achieves the best accuracy on 5-fold cross validation (85.81%), while the radial and linear kernels achieve 84.19%. Testing in a blind data set, the polynomial and radial kernels achieve an accuracy of 90.00%, while the linear model achieves 89.33%. The three models reach higher accuracies than previously described methods. A standalone version of CS-AMPPred is available for download at <http://sourceforge.net/projects/csamppred/> and runs on any Linux machine.
Conflict of interest statement
Figures
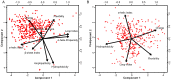
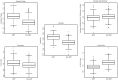
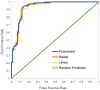
Similar articles
-
deep-AMPpred: A Deep Learning Method for Identifying Antimicrobial Peptides and Their Functional Activities.J Chem Inf Model. 2025 Jan 27;65(2):997-1008. doi: 10.1021/acs.jcim.4c01913. Epub 2025 Jan 10. J Chem Inf Model. 2025. PMID: 39792442
-
Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou's general PseAAC.Sci Rep. 2017 Feb 13;7:42362. doi: 10.1038/srep42362. Sci Rep. 2017. PMID: 28205576 Free PMC article.
-
Ensemble-AMPPred: Robust AMP Prediction and Recognition Using the Ensemble Learning Method with a New Hybrid Feature for Differentiating AMPs.Genes (Basel). 2021 Jan 21;12(2):137. doi: 10.3390/genes12020137. Genes (Basel). 2021. PMID: 33494403 Free PMC article.
-
Peptide Design Principles for Antimicrobial Applications.J Mol Biol. 2019 Aug 23;431(18):3547-3567. doi: 10.1016/j.jmb.2018.12.015. Epub 2019 Jan 3. J Mol Biol. 2019. PMID: 30611750 Review.
-
Antimicrobial peptides: biochemical determinants of activity and biophysical techniques of elucidating their functionality.World J Microbiol Biotechnol. 2018 Apr 12;34(4):62. doi: 10.1007/s11274-018-2444-5. World J Microbiol Biotechnol. 2018. PMID: 29651655 Review.
Cited by
-
Machine learning tools for peptide bioactivity evaluation - Implications for cell culture media optimization and the broader cultivated meat industry.Curr Res Food Sci. 2024 Sep 16;9:100842. doi: 10.1016/j.crfs.2024.100842. eCollection 2024. Curr Res Food Sci. 2024. PMID: 39435450 Free PMC article.
-
Deep Learning for Novel Antimicrobial Peptide Design.Biomolecules. 2021 Mar 22;11(3):471. doi: 10.3390/biom11030471. Biomolecules. 2021. PMID: 33810011 Free PMC article.
-
Antimicrobial potential of a ponericin-like peptide isolated from Bombyx mori L. hemolymph in response to Pseudomonas aeruginosa infection.Sci Rep. 2022 Sep 15;12(1):15493. doi: 10.1038/s41598-022-19450-8. Sci Rep. 2022. PMID: 36109567 Free PMC article.
-
De Novo Design and In Vitro Testing of Antimicrobial Peptides against Gram-Negative Bacteria.Pharmaceuticals (Basel). 2019 Jun 3;12(2):82. doi: 10.3390/ph12020082. Pharmaceuticals (Basel). 2019. PMID: 31163671 Free PMC article.
-
AGRAMP: machine learning models for predicting antimicrobial peptides against phytopathogenic bacteria.Front Microbiol. 2024 Mar 7;15:1304044. doi: 10.3389/fmicb.2024.1304044. eCollection 2024. Front Microbiol. 2024. PMID: 38516021 Free PMC article.
References
-
- Franco OL (2011) Peptide promiscuity: an evolutionary concept for plant defense. FEBS Lett 585 (7): 995–1000. - PubMed
-
- Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3 (3): 238–250. - PubMed
-
- Porto WF, Souza VA, Nolasco DO, Franco OL (2012) In silico identification of novel hevein-like peptide precursors. Peptides 38: 127–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

