Categorical spectral analysis of periodicity in human and viral genomes
- PMID: 23241388
- PMCID: PMC3561982
- DOI: 10.1093/nar/gks1261
Categorical spectral analysis of periodicity in human and viral genomes
Abstract
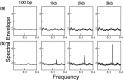
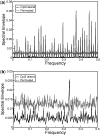
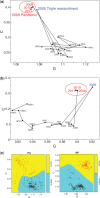
Periodicity in nucleotide sequences arises from regular repeating patterns which may reflect important structure and function. Although a three-base periodicity in coding regions has been known for some time and has provided the basis for powerful gene prediction algorithms, its origins are still not fully understood. Here, we show that, contrary to common belief, amino acid (AA) bias and codon usage bias are insufficient to create base-3 periodicity. This article applies the rigorous method of spectral envelope to systematically characterize the contributions of codon bias, AA bias and protein structural motifs to the three-base periodicity of coding sequences. The method is also used to classify CpG islands in the human genome. In addition, we show how spectral envelope can be used to trace the evolution of viral genomes and monitor global sequence changes without having to align to previously known genomes. This approach also detects reassortment events, such as those that led to the 2009 pandemic H1N1 virus.
Figures
Similar articles
-
Analysis of codon usage preference in hemagglutinin genes of the swine-origin influenza A (H1N1) virus.J Microbiol Immunol Infect. 2016 Aug;49(4):477-86. doi: 10.1016/j.jmii.2014.08.011. Epub 2014 Oct 31. J Microbiol Immunol Infect. 2016. PMID: 25442859
-
Codon and amino acid usage in retroviral genomes is consistent with virus-specific nucleotide pressure.AIDS Res Hum Retroviruses. 2002 Jan 20;18(2):133-41. doi: 10.1089/08892220252779674. AIDS Res Hum Retroviruses. 2002. PMID: 11839146
-
Prediction of directional changes of influenza A virus genome sequences with emphasis on pandemic H1N1/09 as a model case.DNA Res. 2011 Apr;18(2):125-36. doi: 10.1093/dnares/dsr005. Epub 2011 Mar 28. DNA Res. 2011. PMID: 21444341 Free PMC article.
-
On the origin of three base periodicity in genomes.Biosystems. 2012 Mar;107(3):142-4. doi: 10.1016/j.biosystems.2011.11.006. Epub 2011 Nov 12. Biosystems. 2012. PMID: 22100873
-
Attenuation of Human Respiratory Viruses by Synonymous Genome Recoding.Front Immunol. 2019 Jun 4;10:1250. doi: 10.3389/fimmu.2019.01250. eCollection 2019. Front Immunol. 2019. PMID: 31231383 Free PMC article. Review.
Cited by
-
An Adaptive Mapping Method Using Spectral Envelope Approach for DNA Spectral Analysis.Entropy (Basel). 2022 Jul 15;24(7):978. doi: 10.3390/e24070978. Entropy (Basel). 2022. PMID: 35885202 Free PMC article.
-
A degeneration-reducing criterion for optimal digital mapping of genetic codes.Comput Struct Biotechnol J. 2019 Mar 19;17:406-414. doi: 10.1016/j.csbj.2019.03.007. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 30984363 Free PMC article.
-
Identification of a circular code periodicity in the bacterial ribosome: origin of codon periodicity in genes?RNA Biol. 2020 Apr;17(4):571-583. doi: 10.1080/15476286.2020.1719311. Epub 2020 Feb 11. RNA Biol. 2020. PMID: 31960748 Free PMC article.
-
The Heterogeneous Landscape and Early Evolution of Pathogen-Associated CpG Dinucleotides in SARS-CoV-2.SSRN [Preprint]. 2020 May 27:3611280. doi: 10.2139/ssrn.3611280. SSRN. 2020. Update in: Mol Biol Evol. 2021 May 19;38(6):2428-2445. doi: 10.1093/molbev/msab036. PMID: 32714120 Free PMC article. Updated. Preprint.
-
The Heterogeneous Landscape and Early Evolution of Pathogen-Associated CpG Dinucleotides in SARS-CoV-2.Mol Biol Evol. 2021 May 19;38(6):2428-2445. doi: 10.1093/molbev/msab036. Mol Biol Evol. 2021. PMID: 33555346 Free PMC article.
References
-
- Shepherd JCW. Periodic correlations in DNA sequences and evidence suggesting their evolutionary origin in a comma-less genetic code. J. Mol. Evol. 1981;17:94–102. - PubMed
-
- Trifonov EN. Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16 S rRNA nucleotide sequences. J. Mol. Biol. 1987;194:643–652. - PubMed

