Distinct mutational behaviors differentiate short tandem repeats from microsatellites in the human genome
- PMID: 23241442
- PMCID: PMC3622297
- DOI: 10.1093/gbe/evs116
Distinct mutational behaviors differentiate short tandem repeats from microsatellites in the human genome
Abstract
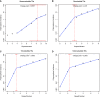
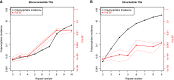
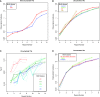
A tandem repeat's (TR) propensity to mutate increases with repeat number, and can become very pronounced beyond a critical boundary, transforming it into a microsatellite (MS). However, a clear understanding of the mutational behavior of different TR classes and motifs and related mechanisms is lacking, as is a consensus on the existence of a boundary separating short TRs (STRs) from MSs. This hinders our understanding of MSs' mutational properties and their effective use as genetic markers. Using indel calls for 179 individuals from 1000 Genomes Pilot-1 Project, we determined polymorphism incidence for four major TR classes, and formalized its varying relationship with repeat number using segmented regression. We observed a biphasic regime with a transition from a faster to a slower exponential growth at 9, 5, 4, and 4 repeats for mono-, di-, tri-, and tetranucleotide TRs, respectively. We used an in vitro mutagenesis assay to evaluate the contribution of strand slippage errors to mutability. STRs and MSs differ in their absolute polymorphism levels, but more importantly in their rates of mutability growth. Although strand slippage is a major factor driving mononucleotide polymorphism incidence, dinucleotide polymorphism incidence is greater than that expected due to strand slippage alone, indicating that additional cellular factors might be driving dinucleotide mutability in the human genome. Leveraging on hundreds of human genomes, we present the first comprehensive, genome-wide analysis of TR mutational behavior, encompassing several motif sizes and compositions.
Figures

References
-
- Bell GI, Jurka J. The length distribution of perfect dimer repetitive DNA is consistent with its evolution by an unbiased single-step mutation process. J Mol Evol. 1997;44:414–421. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

