Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer
- PMID: 23241446
- PMCID: PMC3534625
- DOI: 10.1186/1745-6150-7-46
Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer
Abstract
Background: Collections of Clusters of Orthologous Genes (COGs) provide indispensable tools for comparative genomic analysis, evolutionary reconstruction and functional annotation of new genomes. Initially, COGs were made for all complete genomes of cellular life forms that were available at the time. However, with the accumulation of thousands of complete genomes, construction of a comprehensive COG set has become extremely computationally demanding and prone to error propagation, necessitating the switch to taxon-specific COG collections. Previously, we reported the collection of COGs for 41 genomes of Archaea (arCOGs). Here we present a major update of the arCOGs and describe evolutionary reconstructions to reveal general trends in the evolution of Archaea.
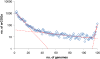
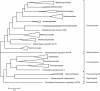
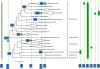
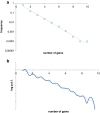
Results: The updated version of the arCOG database incorporates 91% of the pangenome of 120 archaea (251,032 protein-coding genes altogether) into 10,335 arCOGs. Using this new set of arCOGs, we performed maximum likelihood reconstruction of the genome content of archaeal ancestral forms and gene gain and loss events in archaeal evolution. This reconstruction shows that the last Common Ancestor of the extant Archaea was an organism of greater complexity than most of the extant archaea, probably with over 2,500 protein-coding genes. The subsequent evolution of almost all archaeal lineages was apparently dominated by gene loss resulting in genome streamlining. Overall, in the evolution of Archaea as well as a representative set of bacteria that was similarly analyzed for comparison, gene losses are estimated to outnumber gene gains at least 4 to 1. Analysis of specific patterns of gene gain in Archaea shows that, although some groups, in particular Halobacteria, acquire substantially more genes than others, on the whole, gene exchange between major groups of Archaea appears to be largely random, with no major 'highways' of horizontal gene transfer.
Conclusions: The updated collection of arCOGs is expected to become a key resource for comparative genomics, evolutionary reconstruction and functional annotation of new archaeal genomes. Given that, in spite of the major increase in the number of genomes, the conserved core of archaeal genes appears to be stabilizing, the major evolutionary trends revealed here have a chance to stand the test of time.
Reviewers: This article was reviewed by (for complete reviews see the Reviewers' Reports section): Dr. PLG, Prof. PF, Dr. PL (nominated by Prof. JPG).
Figures


Similar articles
-
Archaeal Clusters of Orthologous Genes (arCOGs): An Update and Application for Analysis of Shared Features between Thermococcales, Methanococcales, and Methanobacteriales.Life (Basel). 2015 Mar 10;5(1):818-40. doi: 10.3390/life5010818. Life (Basel). 2015. PMID: 25764277 Free PMC article.
-
Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea.Biol Direct. 2007 Nov 27;2:33. doi: 10.1186/1745-6150-2-33. Biol Direct. 2007. PMID: 18042280 Free PMC article.
-
Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes.BMC Evol Biol. 2003 Jan 6;3:2. doi: 10.1186/1471-2148-3-2. Epub 2003 Jan 6. BMC Evol Biol. 2003. PMID: 12515582 Free PMC article.
-
Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review.
-
Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier?Philos Trans R Soc Lond B Biol Sci. 2015 Sep 26;370(1678):20140333. doi: 10.1098/rstb.2014.0333. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26323764 Free PMC article. Review.
Cited by
-
Rooting the domain archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota.Genome Biol Evol. 2014 Dec 19;7(1):191-204. doi: 10.1093/gbe/evu274. Genome Biol Evol. 2014. PMID: 25527841 Free PMC article.
-
Archaeal Clusters of Orthologous Genes (arCOGs): An Update and Application for Analysis of Shared Features between Thermococcales, Methanococcales, and Methanobacteriales.Life (Basel). 2015 Mar 10;5(1):818-40. doi: 10.3390/life5010818. Life (Basel). 2015. PMID: 25764277 Free PMC article.
-
The Turbulent Network Dynamics of Microbial Evolution and the Statistical Tree of Life.J Mol Evol. 2015 Jun;80(5-6):244-50. doi: 10.1007/s00239-015-9679-7. Epub 2015 Apr 18. J Mol Evol. 2015. PMID: 25894542 Free PMC article. Review.
-
Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions.F1000Res. 2016 Jul 25;5:F1000 Faculty Rev-1805. doi: 10.12688/f1000research.8737.1. eCollection 2016. F1000Res. 2016. PMID: 27508073 Free PMC article. Review.
-
Dissecting the Repertoire of DNA-Binding Transcription Factors of the Archaeon Pyrococcus furiosus DSM 3638.Life (Basel). 2018 Sep 21;8(4):40. doi: 10.3390/life8040040. Life (Basel). 2018. PMID: 30248960 Free PMC article.
References
-
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24(11):539–551. - PubMed
-
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. - PubMed
-
- Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20(11):544–549. - PubMed
-
- Ohno S. Evolution by gene duplication. Berlin-Heidelberg-New York: Springer-Verlag; 1970.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

