Niche-modulated and niche-modulating genes in bone marrow cells
- PMID: 23241658
- PMCID: PMC3542477
- DOI: 10.1038/bcj.2012.42
Niche-modulated and niche-modulating genes in bone marrow cells
Abstract
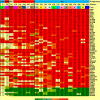
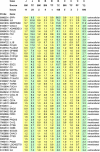
Bone marrow (BM) cells depend on their niche for growth and survival. However, the genes modulated by niche stimuli have not been discriminated yet. For this purpose, we investigated BM aspirations from patients with various hematological malignancies. Each aspirate was fractionated, and the various samples were fixed at different time points and analyzed by microarray. Identification of niche-modulated genes relied on sustained change in expression following loss of niche regulation. Compared with the reference ('authentic') samples, which were fixed immediately following aspiration, the BM samples fixed after longer stay out-of-niche acquired numerous changes in gene-expression profile (GEP). The overall genes modulated included a common subset of functionally diverse genes displaying prompt and sustained 'switch' in expression irrespective of the tumor type. Interestingly, the 'switch' in GEP was reversible and turned 'off-and-on' again in culture conditions, resuming cell-cell-matrix contact versus respread into suspension, respectively. Moreover, the resuming of contact prolonged the survival of tumor cells out-of-niche, and the regression of the 'contactless switch' was followed by induction of a new set of genes, this time mainly encoding extracellular proteins including angiogenic factors and extracellular matrix proteins. Our data set, being unique in authentic expression design, uncovered niche-modulated and niche-modulating genes capable of controlling homing, expansion and angiogenesis.
Figures



References
-
- Yaccoby S, Barlogie B, Epstein J. Primary myeloma cells growing in SCID-hu mice: a model for studying the Biology and treatment of myeloma and its manifestations. Blood. 1998;92:2908–2913. - PubMed
-
- Nefedova Y, Landowski TH, Dalton WS. Bone marrow stromal-derived soluble factors and direct cell contact contribute to de novo drug resistance of myeloma cells by distinct mechanisms. Leukemia. 2003;17:1175–1182. - PubMed
-
- Kohlmann A, Bullinger L, Thiede C, Schaich M, Schnittger S, Döhner K, et al. Gene expression profiling in AML with normal karyotype can predict mutations for molecular markers and allows novel insights into perturbed biological pathways. Leukemia. 2010;24:1216–1220. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

