The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation
- PMID: 23242216
- PMCID: PMC3786872
- DOI: 10.1038/ncb2638
The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation
Abstract
The generation of research-quality, clinically relevant cell types in vitro from human pluripotent stem cells requires a detailed understanding of the equivalent human cell types. Here we analysed 134 human embryonic and fetal samples from 6 to 20 developmental weeks and identified the stages at which cKIT(+) primordial germ cells (PGCs), the precursors of gametes, undergo whole-genome epigenetic reprogramming with global depletion of 5mC, H3K27me3 and H2A.Z, and the time at which imprint erasure is initiated and 5hmC is present. Using five alternative in vitro differentiation strategies combined with single-cell microfluidic analysis and a bona fide human cKIT(+) PGC signature, we show the stage of cKIT(+) PGC formation in the first 16 days of differentiation. Taken together, our study creates a resource of human germ line ontogeny that is essential for future studies aimed at in vitro differentiation and unveiling the mechanisms necessary to pass human DNA from one generation to the next.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

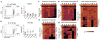




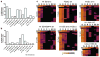

Comment in
-
Humans put their eggs in more than one basket.Nat Cell Biol. 2013 Jan;15(1):13-5. doi: 10.1038/ncb2665. Nat Cell Biol. 2013. PMID: 23263370 Free PMC article.
-
Forget the Parents: Epigenetic Reprogramming in Human Germ Cells.Cell. 2015 Jun 4;161(6):1248-51. doi: 10.1016/j.cell.2015.05.039. Cell. 2015. PMID: 26046435
References
-
- Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–84. - PubMed
-
- Hayashi K, et al. Offspring from Oocytes Derived from in Vitro Primordial Germ Cell-Like Cells in Mice. Science. 2012 Oct 4; [Epub ahead of print] - PubMed
-
- Reijo R, et al. Mouse autosomal homolog of DAZ, a candidate male sterility gene in humans, is expressed in male germ cells before and after puberty. Genomics. 1996;35:346–352. - PubMed
-
- Gaskell TL, Esnal A, Robinson LLL, Anderson RA, Saunders PTK. Immunohistochemical profiling of germ cells within the human fetal testis: identification of three subpopulations. Biol Reprod. 2004;71:2012–2021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

