Diversity in glycosaminoglycan binding amongst hMPV G protein lineages
- PMID: 23242371
- PMCID: PMC3528290
- DOI: 10.3390/v4123785
Diversity in glycosaminoglycan binding amongst hMPV G protein lineages
Abstract
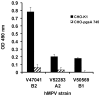
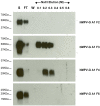
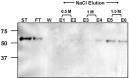
We have previously shown that hMPV G protein (B2 lineage) interacts with cellular glycosaminoglycans (GAGs). In this study we examined subtypes A1, A2 and B1 for this interaction. GAG-dependent infectivity of available hMPV strains was demonstrated using GAG-deficient cells and heparin competition. We expressed the G protein ectodomains from all strains and analysed these by heparin affinity chromatography. In contrast to the B2 lineage, neither the A2 or B1 G proteins bound to heparin. Sequence analysis of these strains indicated that although there was some homology with the B2 heparin-binding domains, there were less positively charged residues, providing a likely explanation for the lack of binding. Although sequence analysis did not demonstrate well defined positively charged domains in G protein of the A1 strain, this protein was able to bind heparin, albeit with a lower affinity than G protein of the B2 strain. These results indicate diversity in GAG interactions between G proteins of different lineages and suggest that the GAG-dependency of all strains may be mediated by interaction with an alternative surface protein, most probably the conserved fusion (F) protein. Analysis of both native and recombinant F protein confirmed that F protein binds heparin, supporting this conclusion.
Figures




References
-
- Boivin G., Abed Y., Pelletier G., Ruel L., Moisan D., Cote S., Peret T.C., Erdman D.D., Anderson L.J. Virological features and clinical manifestations associated with human metapneumovirus: A new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 2002;186:1330–1334. doi: 10.1086/344319. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

