Differential evolution and neofunctionalization of snake venom metalloprotease domains
- PMID: 23242553
- PMCID: PMC3591658
- DOI: 10.1074/mcp.M112.023135
Differential evolution and neofunctionalization of snake venom metalloprotease domains
Erratum in
- Mol Cell Proteomics. 2013 May;12(5):1488. Yang, Dary C [corrected to Yang, Daryl C]
Abstract
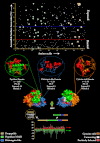
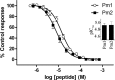
Snake venom metalloproteases (SVMP) are composed of five domains: signal peptide, propeptide, metalloprotease, disintegrin, and cysteine-rich. Secreted toxins are typically combinatorial variations of the latter three domains. The SVMP-encoding genes of Psammophis mossambicus venom are unique in containing only the signal and propeptide domains. We show that the Psammophis SVMP propeptide evolves rapidly and is subject to a high degree of positive selection. Unlike Psammophis, some species of Echis express both the typical multidomain and the unusual monodomain (propeptide only) SVMP, with the result that a lower level of variation is exerted upon the latter. We showed that most mutations in the multidomain Echis SVMP occurred in the protease domain responsible for proteolytic and hemorrhagic activities. The cysteine-rich and disintegrin-like domains, which are putatively responsible for making the P-III SVMPs more potent than the P-I and P-II forms, accumulate the remaining variation. Thus, the binding sites on the molecule's surface are evolving rapidly whereas the core remains relatively conserved. Bioassays conducted on two post-translationally cleaved novel proline-rich peptides from the P. mossambicus propeptide domain showed them to have been neofunctionalized for specific inhibition of mammalian a7 neuronal nicotinic acetylcholine receptors. We show that the proline rich postsynaptic specific neurotoxic peptides from Azemiops feae are the result of convergent evolution within the precursor region of the C-type natriuretic peptide instead of the SVMP. The results of this study reinforce the value of studying obscure venoms for biodiscovery of novel investigational ligands.
Figures




References
-
- Fry B. G., Wüster W. (2004) Assembling an arsenal: origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 21, 870–883 - PubMed
-
- Fry B. G., Scheib H., van der Weerd L., Young B., McNaughtan J., Ryan Ramjan S. F., Vidal N., Poelmann R. E., Norman J. A. (2008) Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteomics 7, 215–246 - PubMed
-
- Weldon C. L., Mackessy S. P. (2012) Alsophinase, a new P-III metalloproteinase with alpha-fibrinogenolytic and hemorrhagic activity from the venom of the rear-fanged Puerto Rican Racer Alsophis portoricensis (Serpentes: Dipsadidae). Biochimie 94, 1189–1198 - PubMed
-
- Peichoto M. E., Leme A. F., Pauletti B. A., Batista I. C., Mackessy S. P., Acosta O., Santoro M. L. (2010) Autolysis at the disintegrin domain of patagonfibrase, a metalloproteinase from Philodryas patagoniensis (Patagonia Green Racer; Dipsadidae) venom. Biochim. Biophys. Acta 1804, 1937–1942 - PubMed
-
- Fry B. G., Scheib H., de L. M. J. d. A. I., Silva D. A., Casewell N. R. (2012) Novel transcripts in the maxillary venom glands of advanced snakes. Toxicon 59, 696–708 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

