Arginine deiminase inhibits Porphyromonas gingivalis surface attachment
- PMID: 23242802
- PMCID: PMC3709564
- DOI: 10.1099/mic.0.062695-0
Arginine deiminase inhibits Porphyromonas gingivalis surface attachment
Abstract
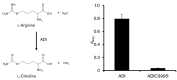
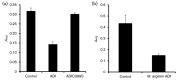
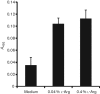
The oral cavity is host to a complex microbial community whose maintenance depends on an array of cell-to-cell interactions and communication networks, with little known regarding the nature of the signals or mechanisms by which they are sensed and transmitted. Determining the signals that control attachment, biofilm development and outgrowth of oral pathogens is fundamental to understanding pathogenic biofilm development. We have previously identified a secreted arginine deiminase (ADI) produced by Streptococcus intermedius that inhibited biofilm development of the commensal pathogen Porphyromonas gingivalis through downregulation of genes encoding the major (fimA) and minor (mfa1) fimbriae, both of which are required for proper biofilm development. Here we report that this inhibitory effect is dependent on enzymic activity. We have successfully cloned, expressed and defined the conditions to ensure that ADI from S. intermedius is enzymically active. Along with the cloning of the wild-type allele, we have created a catalytic mutant (ADIC399S), in which the resulting protein is not able to catalyse the hydrolysis of l-arginine to l-citrulline. P. gingivalis is insensitive to the ADIC399S catalytic mutant, demonstrating that enzymic activity is required for the effects of ADI on biofilm formation. Biofilm formation is absent under l-arginine-deplete conditions, and can be recovered by the addition of the amino acid. Taken together, the results indicate that arginine is an important signal that directs biofilm formation by this anaerobe. Based on our findings, we postulate that ADI functions to reduce arginine levels and, by a yet to be identified mechanism, signals P. gingivalis to alter biofilm development. ADI release from the streptococcal cell and its cross-genera effects are important findings in understanding the nature of inter-bacterial signalling and biofilm-mediated diseases of the oral cavity.
Figures


Similar articles
-
A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development.Microbiology (Reading). 2010 Nov;156(Pt 11):3469-3477. doi: 10.1099/mic.0.042671-0. Epub 2010 Aug 12. Microbiology (Reading). 2010. PMID: 20705665 Free PMC article.
-
Role of arginine deiminase of Streptococcus cristatus in Porphyromonas gingivalis colonization.Antimicrob Agents Chemother. 2010 Nov;54(11):4694-8. doi: 10.1128/AAC.00284-10. Epub 2010 Jul 26. Antimicrob Agents Chemother. 2010. PMID: 20660674 Free PMC article.
-
Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation.Infect Immun. 2006 Oct;74(10):5756-62. doi: 10.1128/IAI.00813-06. Infect Immun. 2006. PMID: 16988253 Free PMC article.
-
Dual lifestyle of Porphyromonas gingivalis in biofilm and gingival cells.Microb Pathog. 2016 May;94:42-7. doi: 10.1016/j.micpath.2015.10.003. Epub 2015 Oct 9. Microb Pathog. 2016. PMID: 26456558 Review.
-
Microbial interactions in building of communities.Mol Oral Microbiol. 2013 Apr;28(2):83-101. doi: 10.1111/omi.12012. Epub 2012 Dec 17. Mol Oral Microbiol. 2013. PMID: 23253299 Free PMC article. Review.
Cited by
-
Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis.Sci Rep. 2017 May 3;7(1):1413. doi: 10.1038/s41598-017-01551-4. Sci Rep. 2017. PMID: 28469253 Free PMC article.
-
New approaches to combat Porphyromonas gingivalis biofilms.J Oral Microbiol. 2017 Mar 15;9(1):1300366. doi: 10.1080/20002297.2017.1300366. eCollection 2017. J Oral Microbiol. 2017. PMID: 28473880 Free PMC article. Review.
-
Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381.NPJ Biofilms Microbiomes. 2019 Feb 7;5(1):7. doi: 10.1038/s41522-019-0081-x. NPJ Biofilms Microbiomes. 2019. PMID: 32029738 Free PMC article.
-
Genes Contributing to Porphyromonas gingivalis Fitness in Abscess and Epithelial Cell Colonization Environments.Front Cell Infect Microbiol. 2017 Aug 28;7:378. doi: 10.3389/fcimb.2017.00378. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28900609 Free PMC article.
-
Gemella haemolysans inhibits the growth of the periodontal pathogen Porphyromonas gingivalis.Sci Rep. 2021 Jun 3;11(1):11742. doi: 10.1038/s41598-021-91267-3. Sci Rep. 2021. PMID: 34083694 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

