The adaptation of temperate bacteriophages to their host genomes
- PMID: 23243039
- PMCID: PMC3603311
- DOI: 10.1093/molbev/mss279
The adaptation of temperate bacteriophages to their host genomes
Abstract
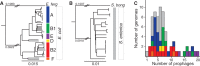
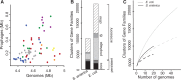
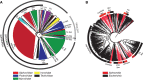
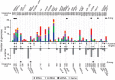
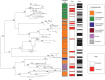
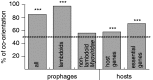
Rapid turnover of mobile elements drives the plasticity of bacterial genomes. Integrated bacteriophages (prophages) encode host-adaptive traits and represent a sizable fraction of bacterial chromosomes. We hypothesized that natural selection shapes prophage integration patterns relative to the host genome organization. We tested this idea by detecting and studying 500 prophages of 69 strains of Escherichia and Salmonella. Phage integrases often target not only conserved genes but also intergenic positions, suggesting purifying selection for integration sites. Furthermore, most integration hotspots are conserved between the two host genera. Integration sites seem also selected at the large chromosomal scale, as they are nonrandomly organized in terms of the origin-terminus axis and the macrodomain structure. The genes of lambdoid prophages are systematically co-oriented with the bacterial replication fork and display the host high frequency of polarized FtsK-orienting polar sequences motifs required for chromosome segregation. matS motifs are strongly avoided by prophages suggesting counter selection of motifs disrupting macrodomains. These results show how natural selection for seamless integration of prophages in the chromosome shapes the evolution of the bacterium and the phage. First, integration sites are highly conserved for many millions of years favoring lysogeny over the lytic cycle for temperate phages. Second, the global distribution of prophages is intimately associated with the chromosome structure and the patterns of gene expression. Third, the phage endures selection for DNA motifs that pertain exclusively to the biology of the prophage in the bacterial chromosome. Understanding prophage genetic adaptation sheds new lights on the coexistence of horizontal transfer and organized bacterial genomes.
Figures

References
-
- Abedon ST, Calendar RL. The bacteriophages. New York: Oxford University Press; 2005.
-
- Ackermann HW. Salmonella phages examined in the electron microscope. Methods Mol Biol. 2007;394:213–234. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

