Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia
- PMID: 23243274
- PMCID: PMC3578955
- DOI: 10.1182/blood-2012-09-458265
Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia
Abstract
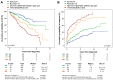
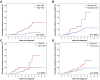
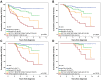
The identification of new genetic lesions in chronic lymphocytic leukemia (CLL) prompts a comprehensive and dynamic prognostic algorithm including gene mutations and chromosomal abnormalities and their changes during clonal evolution. By integrating mutational and cytogenetic analysis in 1274 CLL samples and using both a training-validation and a time-dependent design, 4 CLL subgroups were hierarchically classified: (1) high-risk, harboring TP53 and/or BIRC3 abnormalities (10-year survival: 29%); (2) intermediate-risk, harboring NOTCH1 and/or SF3B1 mutations and/or del11q22-q23 (10-year survival: 37%); (3) low-risk, harboring +12 or a normal genetics (10-year survival: 57%); and (4) very low-risk, harboring del13q14 only, whose 10-year survival (69.3%) did not significantly differ from a matched general population. This integrated mutational and cytogenetic model independently predicted survival, improved CLL prognostication accuracy compared with FISH karyotype (P < .0001), and was externally validated in an independent CLL cohort. Clonal evolution from lower to higher risk implicated the emergence of NOTCH1, SF3B1, and BIRC3 abnormalities in addition to TP53 and 11q22-q23 lesions. By taking into account clonal evolution through time-dependent analysis, the genetic model maintained its prognostic relevance at any time from diagnosis. These findings may have relevant implications for the design of clinical trials aimed at assessing the use of mutational profiling to inform therapeutic decisions.
Figures


References
-
- Dighiero G, Hamblin TJ. Chronic lymphocytic leukaemia. Lancet. 2008;371(9617):1017–1029. - PubMed
-
- Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. - PMC - PubMed
-
- Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. - PubMed
-
- Pospisilova S, Gonzalez D, Malcikova J, et al. ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26(7):1458–1461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

