Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei
- PMID: 23243301
- PMCID: PMC3562093
- DOI: 10.1128/JB.01688-12
Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei
Abstract
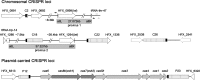
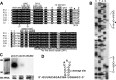
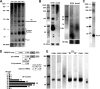
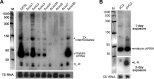
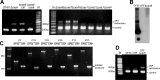
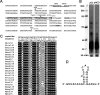
The adaptive immune system comprising CRISPR (clustered regularly interspaced short palindromic repeats) arrays and cas (CRISPR-associated) genes has been discovered in a wide range of bacteria and archaea and has recently attracted comprehensive investigations. However, the subtype I-B CRISPR-Cas system in haloarchaea has been less characterized. Here, we investigated Cas6-mediated RNA processing in Haloferax mediterranei. The Cas6 cleavage site, as well as the CRISPR transcription start site, was experimentally determined, and processing of CRISPR transcripts was detected with a progressively increasing pattern from early log to stationary phase. With genetic approaches, we discovered that the lack of Cas1, Cas3, or Cas4 unexpectedly resulted in a decrease of CRISPR transcripts, while Cas5, Cas6, and Cas7 were found to be essential in stabilizing mature CRISPR RNA (crRNA). Intriguingly, we observed a CRISPR- and Cas3-independent inhibition of a defective provirus, in which the putative Cascade (CRISPR-associated complex for antiviral defense) proteins (Cas5, Cas6, Cas7, and Cas8b) were indispensably required. A sequence carried by a proviral transcript was found to be homologous to the CRISPR repeat RNA and vulnerable to Cas6-mediated cleavage, implying a distinct interference mechanism that may account for this unusual inhibition. These results provide fundamental information for the subtype I-B CRISPR-Cas system in halophilic archaea and suggest diversified mechanisms and multiple physiological functions for the CRISPR-Cas system.
Figures
Similar articles
-
A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii.J Biol Chem. 2014 Mar 7;289(10):7164-7177. doi: 10.1074/jbc.M113.508184. Epub 2014 Jan 23. J Biol Chem. 2014. PMID: 24459147 Free PMC article.
-
Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus.RNA Biol. 2013 May;10(5):738-48. doi: 10.4161/rna.23798. Epub 2013 Feb 7. RNA Biol. 2013. PMID: 23392249 Free PMC article.
-
An active immune defense with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein.J Biol Chem. 2015 Feb 13;290(7):4192-201. doi: 10.1074/jbc.M114.617506. Epub 2014 Dec 15. J Biol Chem. 2015. PMID: 25512373 Free PMC article.
-
Requirements for a successful defence reaction by the CRISPR-Cas subtype I-B system.Biochem Soc Trans. 2013 Dec;41(6):1444-8. doi: 10.1042/BST20130098. Biochem Soc Trans. 2013. PMID: 24256235 Review.
-
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review.
Cited by
-
Primed adaptation tolerates extensive structural and size variations of the CRISPR RNA guide in Haloarcula hispanica.Nucleic Acids Res. 2019 Jun 20;47(11):5880-5891. doi: 10.1093/nar/gkz244. Nucleic Acids Res. 2019. PMID: 30957847 Free PMC article.
-
Molecular characterization of pHRDV1, a new virus-like mobile genetic element closely related to pleomorphic viruses in haloarchaea.Extremophiles. 2014 Mar;18(2):195-206. doi: 10.1007/s00792-013-0599-4. Epub 2013 Dec 29. Extremophiles. 2014. PMID: 24374718
-
Function of the CRISPR-Cas System of the Human Pathogen Clostridium difficile.mBio. 2015 Sep 1;6(5):e01112-15. doi: 10.1128/mBio.01112-15. mBio. 2015. PMID: 26330515 Free PMC article.
-
CRISPR Arrays Away from cas Genes.CRISPR J. 2020 Dec;3(6):535-549. doi: 10.1089/crispr.2020.0062. CRISPR J. 2020. PMID: 33346707 Free PMC article.
-
The Viral Susceptibility of the Haloferax Species.Viruses. 2022 Jun 20;14(6):1344. doi: 10.3390/v14061344. Viruses. 2022. PMID: 35746816 Free PMC article.
References
-
- Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 - PubMed
-
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407 - PubMed
-
- Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338 - PubMed
-
- Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

