PanG, a new ketopantoate reductase involved in pantothenate synthesis
- PMID: 23243306
- PMCID: PMC3571331
- DOI: 10.1128/JB.01740-12
PanG, a new ketopantoate reductase involved in pantothenate synthesis
Abstract
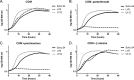
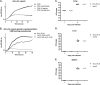
Pantothenate, commonly referred to as vitamin B(5), is an essential molecule in the metabolism of living organisms and forms the core of coenzyme A. Unlike humans, some bacteria and plants are capable of de novo biosynthesis of pantothenate, making this pathway a potential target for drug development. Francisella tularensis subsp. tularensis Schu S4 is a zoonotic bacterial pathogen that is able to synthesize pantothenate but is lacking the known ketopantoate reductase (KPR) genes, panE and ilvC, found in the canonical Escherichia coli pathway. Described herein is a gene encoding a novel KPR, for which we propose the name panG (FTT1388), which is conserved in all sequenced Francisella species and is the sole KPR in Schu S4. Homologs of this KPR are present in other pathogenic bacteria such as Enterococcus faecalis, Coxiella burnetii, and Clostridium difficile. Both the homologous gene from E. faecalis V583 (EF1861) and E. coli panE functionally complemented Francisella novicida lacking any KPR. Furthermore, panG from F. novicida can complement an E. coli KPR double mutant. A Schu S4 ΔpanG strain is a pantothenate auxotroph and was genetically and chemically complemented with panG in trans or with the addition of pantolactone. There was no virulence defect in the Schu S4 ΔpanG strain compared to the wild type in a mouse model of pneumonic tularemia. In summary, we characterized the pantothenate pathway in Francisella novicida and F. tularensis and identified an unknown and previously uncharacterized KPR that can convert 2-dehydropantoate to pantoate, PanG.
Figures

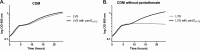
References
-
- Leonardi R, Zhang YM, Rock CO, Jackowski S. 2005. Coenzyme A: back in action. Prog. Lipid Res. 44: 125– 153 - PubMed
-
- Manch JN. 1981. Mapping of a new pan mutation in Escherichia coli K-12. Can. J. Microbiol. 27: 1231– 1233 - PubMed
-
- Elischewski F, Puhler A, Kalinowski J. 1999. Pantothenate production in Escherichia coli K12 by enhanced expression of the panE gene encoding ketopantoate reductase. J. Biotechnol. 75: 135– 146 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

