High-resolution deep sequencing reveals biodiversity, population structure, and persistence of HIV-1 quasispecies within host ecosystems
- PMID: 23244298
- PMCID: PMC3531307
- DOI: 10.1186/1742-4690-9-108
High-resolution deep sequencing reveals biodiversity, population structure, and persistence of HIV-1 quasispecies within host ecosystems
Abstract
Background: Deep sequencing provides the basis for analysis of biodiversity of taxonomically similar organisms in an environment. While extensively applied to microbiome studies, population genetics studies of viruses are limited. To define the scope of HIV-1 population biodiversity within infected individuals, a suite of phylogenetic and population genetic algorithms was applied to HIV-1 envelope hypervariable domain 3 (Env V3) within peripheral blood mononuclear cells from a group of perinatally HIV-1 subtype B infected, therapy-naïve children.
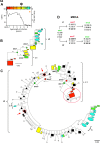
Results: Biodiversity of HIV-1 Env V3 quasispecies ranged from about 70 to 270 unique sequence clusters across individuals. Viral population structure was organized into a limited number of clusters that included the dominant variants combined with multiple clusters of low frequency variants. Next generation viral quasispecies evolved from low frequency variants at earlier time points through multiple non-synonymous changes in lineages within the evolutionary landscape. Minor V3 variants detected as long as four years after infection co-localized in phylogenetic reconstructions with early transmitting viruses or with subsequent plasma virus circulating two years later.
Conclusions: Deep sequencing defines HIV-1 population complexity and structure, reveals the ebb and flow of dominant and rare viral variants in the host ecosystem, and identifies an evolutionary record of low-frequency cell-associated viral V3 variants that persist for years. Bioinformatics pipeline developed for HIV-1 can be applied for biodiversity studies of virome populations in human, animal, or plant ecosystems.
Figures



References
-
- Goodenow M, Huet T, Saurin W, Kwok S, Sninsky J, Wain-Hobson S. HIV-1 isolates are rapidly evolving quasispecies: evidence for viral mixtures and preferred nucleotide substitutions. J Acquir Immune Defic Syndr. 1989;2(4):344–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

