CG methylation
- PMID: 23244310
- PMCID: PMC3566568
- DOI: 10.2217/epi.12.55
CG methylation
Abstract
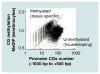
A striking feature of mammalian genomes is the paucity of the CG dinucleotide. There are approximately 20,000 regions termed CpG islands where CGs cluster. This represents 5% of all CGs and 1% of the genome. CpG islands are typically unmethylated and are often promoters for housekeeping genes. The remaining 95% of CG dinucleotides are disposed throughout 99% of the genome and are typically methylated and found in half of all promoters. CG methylation facilitates binding of the C/EBP family of transcription factors, proteins critical for differentiation of many tissues. This allows these proteins to localize in the methylated CG poor regions of the genome where they may produce advantageous changes in gene expression at nearby or more distant regions of the genome. In this review, our growing understanding of the consequences of CG methylation will be surveyed.
Figures

References
-
- Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978;118(1):27–47. - PubMed
-
- Adams RL, McKay EL, Craig LM, Burdon RH. Methylation of mosquito DNA. Biochim Biophys Acta. 1979;563(1):72–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
