Sculpting of DNA at abasic sites by DNA glycosylase homolog mag2
- PMID: 23245849
- PMCID: PMC3545110
- DOI: 10.1016/j.str.2012.11.004
Sculpting of DNA at abasic sites by DNA glycosylase homolog mag2
Abstract
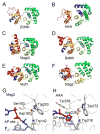
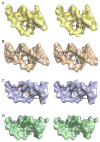
Modifications and loss of bases are frequent types of DNA lesions, often handled by the base excision repair (BER) pathway. BER is initiated by DNA glycosylases, generating abasic (AP) sites that are subsequently cleaved by AP endonucleases, which further pass on nicked DNA to downstream DNA polymerases and ligases. The coordinated handover of cytotoxic intermediates between different BER enzymes is most likely facilitated by the DNA conformation. Here, we present the atomic structure of Schizosaccharomyces pombe Mag2 in complex with DNA to reveal an unexpected structural basis for nonenzymatic AP site recognition with an unflipped AP site. Two surface-exposed loops intercalate and widen the DNA minor groove to generate a DNA conformation previously only found in the mismatch repair MutS-DNA complex. Consequently, the molecular role of Mag2 appears to be AP site recognition and protection, while possibly facilitating damage signaling by structurally sculpting the DNA substrate.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Non-productive DNA damage binding by DNA glycosylase-like protein Mag2 from Schizosaccharomyces pombe.DNA Repair (Amst). 2013 Mar 1;12(3):196-204. doi: 10.1016/j.dnarep.2012.12.001. Epub 2012 Dec 28. DNA Repair (Amst). 2013. PMID: 23273506 Free PMC article.
-
A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe.Nucleic Acids Res. 2004 Sep 27;32(17):5119-25. doi: 10.1093/nar/gkh851. Print 2004. Nucleic Acids Res. 2004. PMID: 15452279 Free PMC article.
-
Interaction of apurinic/apyrimidinic endonuclease 2 (Apn2) with Myh1 DNA glycosylase in fission yeast.DNA Repair (Amst). 2014 Mar;15:1-10. doi: 10.1016/j.dnarep.2014.01.001. Epub 2014 Feb 1. DNA Repair (Amst). 2014. PMID: 24559510 Free PMC article.
-
Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
-
Repair of oxidative DNA damage: mechanisms and functions.Cell Biochem Biophys. 2001;35(2):141-70. doi: 10.1385/CBB:35:2:141. Cell Biochem Biophys. 2001. PMID: 11892789 Review.
Cited by
-
RNA sculpting by the primordial Helix-clasp-Helix-Strand-Loop (HcH-SL) motif enforces chemical recognition enabling diverse KH domain functions.J Biol Chem. 2025 May;301(5):108474. doi: 10.1016/j.jbc.2025.108474. Epub 2025 Apr 2. J Biol Chem. 2025. PMID: 40185232 Free PMC article. Review.
-
A New Family of HEAT-Like Repeat Proteins Lacking a Critical Substrate Recognition Motif Present in Related DNA Glycosylases.PLoS One. 2015 May 15;10(5):e0127733. doi: 10.1371/journal.pone.0127733. eCollection 2015. PLoS One. 2015. PMID: 25978435 Free PMC article.
-
The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions.Nature. 2015 Nov 12;527(7577):254-8. doi: 10.1038/nature15728. Epub 2015 Oct 28. Nature. 2015. PMID: 26524531 Free PMC article.
-
Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair.Prog Biophys Mol Biol. 2015 Mar;117(2-3):182-193. doi: 10.1016/j.pbiomolbio.2014.12.004. Epub 2015 Jan 7. Prog Biophys Mol Biol. 2015. PMID: 25576492 Free PMC article. Review.
-
AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop.Nucleic Acids Res. 2020 Jul 27;48(13):7345-7355. doi: 10.1093/nar/gkaa496. Nucleic Acids Res. 2020. PMID: 32542366 Free PMC article.
References
-
- Berti PJ, McCann JA. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem Rev. 2006;106:506–555. - PubMed
-
- Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. - PubMed
-
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

