Y-family polymerase conformation is a major determinant of fidelity and translesion specificity
- PMID: 23245850
- PMCID: PMC3545038
- DOI: 10.1016/j.str.2012.11.005
Y-family polymerase conformation is a major determinant of fidelity and translesion specificity
Abstract
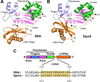
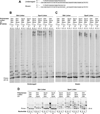
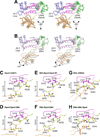
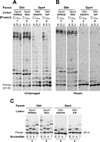
Y-family polymerases help cells tolerate DNA damage by performing translesion synthesis opposite damaged DNA bases, yet they also have a high intrinsic error rate. We constructed chimeras of two closely related Y-family polymerases that display distinctly different activity profiles and found that the polypeptide linker that tethers the catalytic polymerase domain to the C-terminal DNA-binding domain is a major determinant of overall polymerase activity, nucleotide incorporation fidelity, and abasic site-bypass ability. Exchanging just 3 out of the 15 linker residues is sufficient to interconvert the polymerase activities tested. Crystal structures of four chimeras show that the conformation of the protein correlates with the identity of the interdomain linker sequence. Thus, residues that are more than 15 Å away from the active site are able to influence many aspects of polymerase activity by altering the relative orientations of the catalytic and DNA-binding domains.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–970. - PubMed
-
- Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. - PubMed
-
- Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J Biol Chem. 2004;279:32932–32940. - PubMed
-
- DeLucia AM, Chaudhuri S, Potapova O, Grindley ND, Joyce CM. The properties of steric gate mutants reveal different constraints within the active sites of Y-family and A-family DNA polymerases. The Journal of biological chemistry. 2006;281:27286–27291. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

