Rare codons regulate KRas oncogenesis
- PMID: 23246410
- PMCID: PMC3567844
- DOI: 10.1016/j.cub.2012.11.031
Rare codons regulate KRas oncogenesis
Abstract
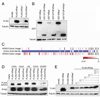
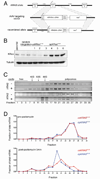
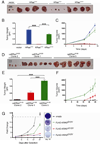
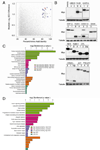
Oncogenic mutations in the small Ras GTPases KRas, HRas, and NRas render the proteins constitutively GTP bound and active, a state that promotes cancer. Ras proteins share ~85% amino acid identity, are activated by and signal through the same proteins, and can exhibit functional redundancy. Nevertheless, manipulating expression or activation of each isoform yields different cellular responses and tumorigenic phenotypes, even when different ras genes are expressed from the same locus. We now report a novel regulatory mechanism hardwired into the very sequence of RAS genes that underlies how such similar proteins impact tumorigenesis differently. Specifically, despite their high sequence similarity, KRAS is poorly translated compared to HRAS due to enrichment in genomically underrepresented or rare codons. Converting rare to common codons increases KRas expression and tumorigenicity to mirror that of HRas. Furthermore, in a genome-wide survey, similar gene pairs with opposing codon bias were identified that not only manifest dichotomous protein expression but also are enriched in key signaling protein classes and pathways. Thus, synonymous nucleotide differences affecting codon usage account for differences between HRas and KRas expression and function and may represent a broader regulation strategy in cell signaling.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Comment in
-
Ras GTPases: codon bias holds KRas down but not out.Curr Biol. 2013 Jan 7;23(1):R17-20. doi: 10.1016/j.cub.2012.11.054. Curr Biol. 2013. PMID: 23305663
References
-
- Quilliam LA, Rebhun JF, Castro AF. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:391–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

