Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches
- PMID: 23246436
- PMCID: PMC3557535
- DOI: 10.1016/j.molcel.2012.11.012
Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches
Abstract
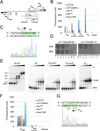
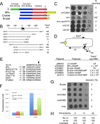
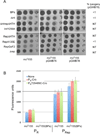
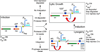
Genetic switches are critical components of developmental circuits. Because temperate bacteriophages are vastly abundant and greatly diverse, they are rich resources for understanding the mechanisms and evolution of switches and the molecular control of genetic circuitry. Here, we describe a new class of small, compact, and simple switches that use site-specific recombination as the key decision point. The phage attachment site attP is located within the phage repressor gene such that chromosomal integration results in removal of a C-terminal tag that destabilizes the virally encoded form of the repressor. Integration thus not only confers prophage stability but also is a requirement for lysogenic establishment. The variety of these self-contained integration-dependent immunity systems in different genomic contexts suggests that these represent ancestral states in switch evolution from which more-complex switches have evolved. They also provide a powerful toolkit for building synthetic biological circuits.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Switches, switches, every where, in any drop we drink.Mol Cell. 2013 Jan 24;49(2):232-3. doi: 10.1016/j.molcel.2013.01.005. Mol Cell. 2013. PMID: 23352244
References
-
- Dodd IB, Shearwin KE, Egan JB. Revisited gene regulation in bacteriophage lambda. Curr Opin Genet Dev. 2005;15:145–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

