A TALEN genome-editing system for generating human stem cell-based disease models
- PMID: 23246482
- PMCID: PMC3570604
- DOI: 10.1016/j.stem.2012.11.011
A TALEN genome-editing system for generating human stem cell-based disease models
Abstract
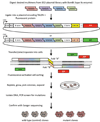
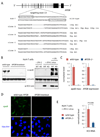
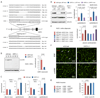
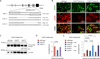
Transcription activator-like effector nucleases (TALENs) are a new class of engineered nucleases that are easier to design to cleave at desired sites in a genome than previous types of nucleases. We report here the use of TALENs to rapidly and efficiently generate mutant alleles of 15 genes in cultured somatic cells or human pluripotent stem cells, the latter for which we differentiated both the targeted lines and isogenic control lines into various metabolic cell types. We demonstrate cell-autonomous phenotypes directly linked to disease-dyslipidemia, insulin resistance, hypoglycemia, lipodystrophy, motor-neuron death, and hepatitis C infection. We found little evidence of TALEN off-target effects, but each clonal line nevertheless harbors a significant number of unique mutations. Given the speed and ease with which we were able to derive and characterize these cell lines, we anticipate TALEN-mediated genome editing of human cells becoming a mainstay for the investigation of human biology and disease.
Copyright © 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
A.M. is a fulltime employee of Roche Pharmaceuticals; the other authors report no relevant conflicts of interest.
Figures

References
-
- Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu. Rev. Genomics Hum. Genet. 2006;7:175–199. - PubMed
-
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. - PubMed
-
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
-
- Brasaemle DL, Subramanian V, Garcia A, Marcinkiewicz A, Rothenberg A. Perilipin A and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 2009;326:15–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 NS066888/NS/NINDS NIH HHS/United States
- U01-HL107440/HL/NHLBI NIH HHS/United States
- T32-HL007604/HL/NHLBI NIH HHS/United States
- K24 DK078772/DK/NIDDK NIH HHS/United States
- R00 HL098364/HL/NHLBI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R00-HL098364/HL/NHLBI NIH HHS/United States
- R01 DK097768/DK/NIDDK NIH HHS/United States
- F32 DK097855/DK/NIDDK NIH HHS/United States
- R01 DK095384/DK/NIDDK NIH HHS/United States
- U01 HL107440/HL/NHLBI NIH HHS/United States
- T32 HL007604/HL/NHLBI NIH HHS/United States
- K08 DK088951/DK/NIDDK NIH HHS/United States
- T32 DK007191/DK/NIDDK NIH HHS/United States
- P01-NS066888/NS/NINDS NIH HHS/United States
- R01-DK097768/DK/NIDDK NIH HHS/United States
- T32-DK007191/DK/NIDDK NIH HHS/United States
- K24-DK078772/DK/NIDDK NIH HHS/United States
- R01 HL118744/HL/NHLBI NIH HHS/United States
- K08-DK088951/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

