Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon
- PMID: 23248290
- PMCID: PMC3538221
- DOI: 10.1073/pnas.1217548110
Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon
Abstract
A revelation of the genomic age has been the contributions of the mobile DNA segments called transposable elements to chromosome structure, function, and evolution in virtually all organisms. Substantial fractions of vertebrate genomes derive from transposable elements, being dominated by retroelements that move via RNA intermediates. Although many of these elements have been inactivated by mutation, several active retroelements remain. Vertebrate genomes also contain substantial quantities and a high diversity of cut-and-paste DNA transposons, but no active representative of this class has been identified in mammals. Here we show that a cut-and-paste element called piggyBat, which has recently invaded the genome of the little brown bat (Myotis lucifugus) and is a member of the piggyBac superfamily, is active in its native form in transposition assays in bat and human cultured cells, as well as in the yeast Saccharomyces cerevisiae. Our study suggests that some DNA transposons are still actively shaping some mammalian genomes and reveals an unprecedented opportunity to study the mechanism, regulation, and genomic impact of cut-and-paste transposition in a natural mammalian host.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

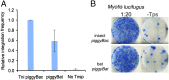
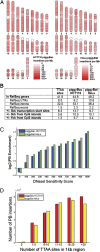

References
-
- Craig NL, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. Washington, DC: ASM Press; 2002.
-
- Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. - PubMed
-
- Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Transposable element in fish. Nature. 1996;383(6595):30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

