Proteasome inhibition rescues clinically significant unstable variants of the mismatch repair protein Msh2
- PMID: 23248292
- PMCID: PMC3538226
- DOI: 10.1073/pnas.1215510110
Proteasome inhibition rescues clinically significant unstable variants of the mismatch repair protein Msh2
Abstract
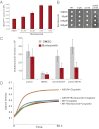
MSH2 is required for DNA mismatch repair recognition in eukaryotes. Deleterious mutations in human MSH2 account for approximately half of the alleles associated with a common hereditary cancer syndrome. Previously, we characterized clinically identified MSH2 missense mutations, using yeast as a model system, and found that the most common cause of defective DNA mismatch repair was low levels of the variant Msh2 proteins. Here, we show that increased protein turnover is responsible for the reduced cellular levels. Increasing gene dosage of more than half of the missense alleles fully restored function. A titration experiment revealed that raising the expression level of one variant to less than wild-type levels restored mismatch repair, suggesting that overexpression is not always required to regain function. We found that the ubiquitin-mediated proteasome degradation pathway is the major mechanism for increased turnover of the Msh2 variants and identified the primary ubiquitin ligase as San1. Deletion of San1 restored protein levels for all but one variant, but did not elevate wild-type Msh2 levels. The unstable variants interacted with San1, whereas wild-type Msh2 did not. Additionally, san1Δ suppressed the mismatch repair defect of unstable variants. Of medical significance, the clinically approved drug Bortezomib partially restored protein levels and mismatch repair function for low-level variants and reversed the resistance to cisplatin, a common chemotherapeutic. Our results provide the foundation for an innovative therapeutic regime for certain mismatch-repair-defective cancers that are refractory to conventional chemotherapies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

