Accurate prediction of secondary metabolite gene clusters in filamentous fungi
- PMID: 23248299
- PMCID: PMC3538241
- DOI: 10.1073/pnas.1205532110
Accurate prediction of secondary metabolite gene clusters in filamentous fungi
Abstract
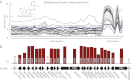
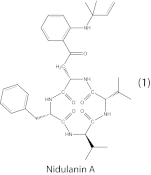
Biosynthetic pathways of secondary metabolites from fungi are currently subject to an intense effort to elucidate the genetic basis for these compounds due to their large potential within pharmaceutics and synthetic biochemistry. The preferred method is methodical gene deletions to identify supporting enzymes for key synthases one cluster at a time. In this study, we design and apply a DNA expression array for Aspergillus nidulans in combination with legacy data to form a comprehensive gene expression compendium. We apply a guilt-by-association-based analysis to predict the extent of the biosynthetic clusters for the 58 synthases active in our set of experimental conditions. A comparison with legacy data shows the method to be accurate in 13 of 16 known clusters and nearly accurate for the remaining 3 clusters. Furthermore, we apply a data clustering approach, which identifies cross-chemistry between physically separate gene clusters (superclusters), and validate this both with legacy data and experimentally by prediction and verification of a supercluster consisting of the synthase AN1242 and the prenyltransferase AN11080, as well as identification of the product compound nidulanin A. We have used A. nidulans for our method development and validation due to the wealth of available biochemical data, but the method can be applied to any fungus with a sequenced and assembled genome, thus supporting further secondary metabolite pathway elucidation in the fungal kingdom.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kelly DE, Krasevec N, Mullins J, Nelson DR. The CYPome (Cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet Biol. 2009;46(Suppl 1):S53–S61. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

