Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants
- PMID: 23248305
- PMCID: PMC3538250
- DOI: 10.1073/pnas.1215376110
Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants
Abstract
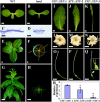
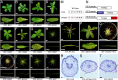
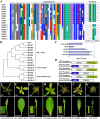
The WUSCHEL related homeobox (WOX) genes play key roles in stem cell maintenance, embryonic patterning, and lateral organ development. WOX genes have been categorized into three clades--ancient, intermediate, and modern/WUS--based on phylogenetic analysis, but a functional basis for this classification has not been established. Using the classical bladeless lam1 mutant of Nicotiana sylvestris as a genetic tool, we examined the function of the Medicago truncatula WOX gene, STENOFOLIA (STF), in controlling leaf blade outgrowth. STF and LAM1 are functional orthologs. We found that the introduction of mutations into the WUS-box of STF (STFm1) reduces its ability to complement the lam1 mutant. Fusion of an exogenous repressor domain to STFm1 restores complementation, whereas fusion of an exogenous activator domain to STFm1 enhances the narrow leaf phenotype. These results indicate that transcriptional repressor activity mediated by the WUS-box of STF acts to promote blade outgrowth. With the exception of WOX7, the WUS-box is conserved in the modern clade WOX genes, but is not found in members of the intermediate or ancient clades. Consistent with this, all members of the modern clade except WOX7 can complement the lam1 mutant when expressed using the STF promoter, but members of the intermediate and ancient clades cannot. Furthermore, we found that fusion of either the WUS-box or an exogenous repressor domain to WOX7 or to members of intermediate and ancient WOX clades results in a gain-of-function ability to complement lam1 blade outgrowth. These results suggest that modern clade WOX genes have evolved for repressor activity through acquisition of the WUS-box.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Haecker A, et al. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131(3):657–668. - PubMed
-
- Ueda M, Zhang Z, Laux T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell. 2011;20(2):264–270. - PubMed
-
- Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446(7137):811–814. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

