Mechanisms of CFTR Folding at the Endoplasmic Reticulum
- PMID: 23248597
- PMCID: PMC3521238
- DOI: 10.3389/fphar.2012.00201
Mechanisms of CFTR Folding at the Endoplasmic Reticulum
Abstract
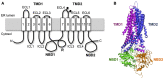
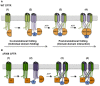
In the past decade much has been learned about how Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) folds and misfolds as the etiologic cause of cystic fibrosis (CF). CFTR folding is complex and hierarchical, takes place in multiple cellular compartments and physical environments, and involves several large networks of folding machineries. Insertion of transmembrane (TM) segments into the endoplasmic reticulum (ER) membrane and tertiary folding of cytosolic domains begin cotranslationally as the nascent polypeptide emerges from the ribosome, whereas posttranslational folding establishes critical domain-domain contacts needed to form a physiologically stable structure. Within the membrane, N- and C-terminal TM helices are sorted into bundles that project from the cytosol to form docking sites for nucleotide binding domains, NBD1 and NBD2, which in turn form a sandwich dimer for ATP binding. While tertiary folding is required for domain assembly, proper domain assembly also reciprocally affects folding of individual domains analogous to a jig-saw puzzle wherein the structure of each interlocking piece influences its neighbors. Superimposed on this process is an elaborate proteostatic network of cellular chaperones and folding machineries that facilitate the timing and coordination of specific folding steps in and across the ER membrane. While the details of this process require further refinement, we finally have a useful framework to understand key folding defect(s) caused by ΔF508 that provides a molecular target(s) for the next generation of CFTR small molecule correctors aimed at the specific defect present in the majority of CF patients.
Keywords: ABC transporter; CFTR; cotranslational folding; cystic fibrosis; membrane protein biogenesis; nucleotide binding domain; protein translocation.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

