Cytochromes b561: ascorbate-mediated trans-membrane electron transport
- PMID: 23249217
- PMCID: PMC3763232
- DOI: 10.1089/ars.2012.5065
Cytochromes b561: ascorbate-mediated trans-membrane electron transport
Abstract
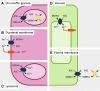
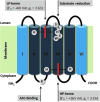
Significance: Cytochromes b561 (CYB561s) constitute a family of trans-membrane (TM), di-heme proteins, occurring in a variety of organs and cell types, in plants and animals, and using ascorbate (ASC) as an electron donor. CYB561s function as monodehydroascorbate reductase, regenerating ASC, and as Fe³⁺-reductases, providing reduced iron for TM transport. A CYB561-core domain is also associated with dopamine β-monooxygenase redox domains (DOMON) in ubiquitous CYBDOM proteins. In plants, CYBDOMs form large protein families. Physiological functions supported by CYB561s and CYBDOMs include stress defense, cell wall modifications, iron metabolism, tumor suppression, and various neurological processes, including memory retention. CYB561s, therefore, significantly broaden our view on the physiological roles of ASC.
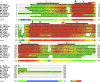
Recent advances: The ubiquitous nature of CYB561s is only recently being recognized. Significant advances have been made through the study of recombinant CYB561s, revealing structural and functional properties of a unique "two-heme four-helix" protein configuration. In addition, the DOMON domains of CYBDOMs are suggested to contain another heme b.
Critical issues: New CYB561 proteins are still being identified, and there is a need to provide an insight and overview on the various roles of these proteins and their structural properties.
Future directions: Mutant studies will reveal in greater detail the mechanisms by which CYB561s and CYBDOMs participate in cell metabolism in plants and animals. Moreover, the availability of efficient heterologous expression systems should allow protein crystallization, more detailed (atomic-level) structural information, and insights into the intra-molecular mechanism of electron transport.
Figures

References
-
- Aravind L. Domon: an ancient extracellular domain in dopamine beta-monooxygenase and other proteins. Trends Biochem Sci. 2001;26:524–526. - PubMed
-
- Asard H. Horemans N. Caubergs RJ. Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett. 1992;306:143–146. - PubMed
-
- Asard H. Terol-Alcayde J. Preger V. Del Favero J. Verelst W. Sparla F. Pérez-Alonso M. Trost P. Arabidopsis thaliana sequence analysis confirms the presence of cyt b-561 in plants: evidence for a novel protein family. Plant Physiol Biochem. 2000;38:905–912.
-
- Askerlund P. Larsson C. Widell S. Cytochromes of plant plasma membranes. Characterization by absorbance difference spectroscopy and redox titration. Physiol Plant. 1989;76:123–134.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

