Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches
- PMID: 23249744
- PMCID: PMC3543082
- DOI: 10.1261/rna.036269.112
Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches
Abstract
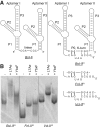
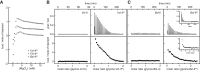
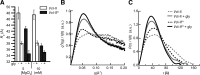
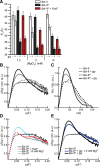
Most known glycine riboswitches have two homologous aptamer domains arranged in tandem and separated by a short linker. The two aptamers associate through reciprocal "quaternary" interactions that have been proposed to result in cooperative glycine binding. Recently, the interaptamer linker was found to form helix P0 with a previously unrecognized segment 5' to the first aptamer domain. P0 was shown to increase glycine affinity, abolish cooperativity, and conform to the K-turn motif consensus. We examine the global thermodynamic and structural role of P0 using isothermal titration calorimetry (ITC) and small-angle X-ray scattering (SAXS), respectively. To evaluate the generality of P0 function, we prepared glycine riboswitch constructs lacking and including P0 from Bacillus subtilis, Fusobacterium nucleatum, and Vibrio cholerae. We find that P0 indeed folds into a K-turn, supports partial pre-folding of all three glycine-free RNAs, and is required for ITC observation of glycine binding under physiologic Mg(2+) concentrations. Except for the unusually small riboswitch from F. nucleatum, the K-turn is needed for maximally compacting the glycine-bound states of the RNAs. Formation of a ribonucleoprotein complex between the B. subtilis or the F. nucleatum RNA constructs and the bacterial K-turn binding protein YbxF promotes additional folding of the free riboswitch, and enhances glycine binding. Consistent with the previously reported loss of cooperativity, P0-containing B. subtilis and V. cholerae tandem aptamers bound no more than one glycine molecule per riboswitch. Our results indicate that the P0 K-turn helps organize the quaternary structure of tandem glycine riboswitches, thereby facilitating ligand binding under physiologic conditions.
Figures
References
-
- Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR 2005. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J Mol Biol 352: 712–722 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
