tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae
- PMID: 23249748
- PMCID: PMC3543094
- DOI: 10.1261/rna.035808.112
tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae
Abstract
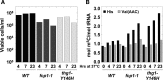
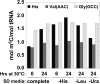
tRNAs are highly modified, each with a unique set of modifications. Several reports suggest that tRNAs are hypomodified or, in some cases, hypermodified under different growth conditions and in certain cancers. We previously demonstrated that yeast strains depleted of tRNA(His) guanylyltransferase accumulate uncharged tRNA(His) lacking the G(-1) residue and subsequently accumulate additional 5-methylcytidine (m(5)C) at residues C(48) and C(50) of tRNA(His), due to the activity of the m(5)C-methyltransferase Trm4. We show here that the increase in tRNA(His) m(5)C levels does not require loss of Thg1, loss of G(-1) of tRNA(His), or cell death but is associated with growth arrest following different stress conditions. We find substantially increased tRNA(His) m(5)C levels after temperature-sensitive strains are grown at nonpermissive temperature, and after wild-type strains are grown to stationary phase, starved for required amino acids, or treated with rapamycin. We observe more modest accumulations of m(5)C in tRNA(His) after starvation for glucose and after starvation for uracil. In virtually all cases examined, the additional m(5)C on tRNA(His) occurs while cells are fully viable, and the increase is neither due to the GCN4 pathway, nor to increased Trm4 levels. Moreover, the increased m(5)C appears specific to tRNA(His), as tRNA(Val(AAC)) and tRNA(Gly(GCC)) have much reduced additional m(5)C during these growth arrest conditions, although they also have C(48) and C(50) and are capable of having increased m(5)C levels. Thus, tRNA(His) m(5)C levels are unusually responsive to yeast growth conditions, although the significance of this additional m(5)C remains unclear.
Figures



References
-
- Agris PF, Koh H, Soll D 1973. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch Biochem Biophys 154: 277–282 - PubMed
-
- Agris PF, Vendeix FA, Graham WD 2007. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 - PubMed
-
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
