Potent microRNA suppression by RNA Pol II-transcribed 'Tough Decoy' inhibitors
- PMID: 23249752
- PMCID: PMC3543086
- DOI: 10.1261/rna.034850.112
Potent microRNA suppression by RNA Pol II-transcribed 'Tough Decoy' inhibitors
Abstract
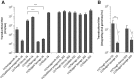
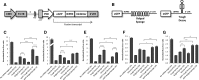
MicroRNAs (miRNAs) are key regulators of gene expression and modulators of diverse biological pathways. Analyses of miRNA function as well as therapeutic managing of miRNAs rely on cellular administration of miRNA inhibitors which may be achieved by the use of viral vehicles. This study explores the miRNA-suppressive capacity of inhibitors expressed intracellularly from lentivirus-derived gene vectors. Superior activity of two decoy-type inhibitors, a "Bulged Sponge" with eight miRNA recognition sites and a hairpin-shaped "Tough Decoy" containing two miRNA recognition sites, is demonstrated in a side-by-side comparison of seven types of miRNA inhibitors transcribed as short RNAs from an RNA Pol III promoter. We find that lentiviral vectors expressing Tough Decoy inhibitors are less vulnerable than Bulged Sponge-encoding vectors to targeting by the cognate miRNA and less prone, therefore, to reductions in transfer efficiency. Importantly, it is demonstrated that Tough Decoy inhibitors retain their miRNA suppression capacity in the context of longer RNA transcripts expressed from an RNA Pol II promoter. Such RNA Pol II-transcribed Tough Decoy inhibitors are new tools in managing of miRNAs and may have potential for temporal and spatial regulation of miRNA activity as well as for therapeutic targeting of miRNAs that are aberrantly expressed in human disease.
Figures




References
-
- Ameres SL, Martinez J, Schroeder R 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 - PubMed
-
- An W, Telesnitsky A 2001. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology 286: 475–482 - PubMed
-
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat Med 13: 613–618 - PubMed
-
- Chang K, Elledge SJ, Hannon GJ 2006. Lessons from Nature: MicroRNA-based shRNA libraries. Nat Methods 3: 707–714 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
