Proteome-wide selected reaction monitoring assays for the human pathogen Streptococcus pyogenes
- PMID: 23250431
- PMCID: PMC3535367
- DOI: 10.1038/ncomms2297
Proteome-wide selected reaction monitoring assays for the human pathogen Streptococcus pyogenes
Abstract
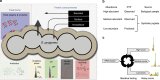
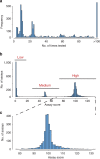
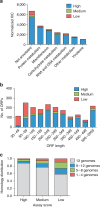
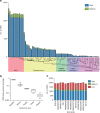
Selected reaction monitoring mass spectrometry (SRM-MS) is a targeted proteomics technology used to identify and quantify proteins with high sensitivity, specificity and high reproducibility. Execution of SRM-MS relies on protein-specific SRM assays, a set of experimental parameters that requires considerable effort to develop. Here we present a proteome-wide SRM assay repository for the gram-positive human pathogen group A Streptococcus. Using a multi-layered approach we generated SRM assays for 10,412 distinct group A Streptococcus peptides followed by extensive testing of the selected reaction monitoring assays in >200 different group A Streptococcus protein pools. Based on the number of SRM assay observations we created a rule-based selected reaction monitoring assay-scoring model to select the most suitable assays per protein for a given cellular compartment and bacterial state. The resource described here represents an important tool for deciphering the group A Streptococcus proteome using selected reaction monitoring and we anticipate that concepts described here can be extended to other pathogens.
Figures

References
-
- Hecker M., Becher D., Fuchs S. & Engelmann S. A proteomic view of cell physiology and virulence of Staphylococcus aureus. Int. J. Med. Microbiol. 300, 76–87 (2010). - PubMed
-
- Poetsch A., Haussmann U. & Burkovski A. Proteomics of corynebacteria: from biotechnology workhorses to pathogens. Proteomics 11, 3244–3255 (2011). - PubMed
-
- Chao T.-C. & Hansmeier N. The current state of microbial proteomics: where we are and where we want to go. Proteomics 12, 638–650 (2012). - PubMed
-
- Cole J. N., Barnett T. C., Nizet V. & Walker M. J. Molecular insight into invasive group. A streptococcal disease. Nat. Rev. Microbiol. 9, 724–736 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

