A measure of the signal-to-noise ratio of microarray samples and studies using gene correlations
- PMID: 23251415
- PMCID: PMC3520972
- DOI: 10.1371/journal.pone.0051013
A measure of the signal-to-noise ratio of microarray samples and studies using gene correlations
Abstract
Background: The quality of gene expression data can vary dramatically from platform to platform, study to study, and sample to sample. As reliable statistical analysis rests on reliable data, determining such quality is of the utmost importance. Quality measures to spot problematic samples exist, but they are platform-specific, and cannot be used to compare studies.
Results: As a proxy for quality, we propose a signal-to-noise ratio for microarray data, the "Signal-to-Noise Applied to Gene Expression Experiments", or SNAGEE. SNAGEE is based on the consistency of gene-gene correlations. We applied SNAGEE to a compendium of 80 large datasets on 37 platforms, for a total of 24,380 samples, and assessed the signal-to-noise ratio of studies and samples. This allowed us to discover serious issues with three studies. We show that signal-to-noise ratios of both studies and samples are linked to the statistical significance of the biological results.
Conclusions: We showed that SNAGEE is an effective way to measure data quality for most types of gene expression studies, and that it often outperforms existing techniques. Furthermore, SNAGEE is platform-independent and does not require raw data files. The SNAGEE R package is available in BioConductor.
Conflict of interest statement
Figures

 are on Affymetrix platforms (GSE5720 and GSE5949),
are on Affymetrix platforms (GSE5720 and GSE5949),  is on NCI dual channel (GSE2003) and
is on NCI dual channel (GSE2003) and  is on Stanford dual channel (GSE7947). c.c. are correlation coefficients.
is on Stanford dual channel (GSE7947). c.c. are correlation coefficients.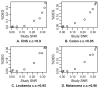
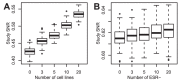
 ) or without (
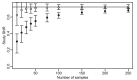
) or without ( ). The error bars are the 95% confidence intervals determined by resampling. The horizontal line is the SNR of the whole data set.
). The error bars are the 95% confidence intervals determined by resampling. The horizontal line is the SNR of the whole data set.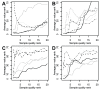
 lowest quality samples is shown. Quality of the samples was determined with SNAGEE (solid line), MDQC (dashed line), GNUSE (dotted line), NUSE (grey dotted line) and RLE (dash-dot line). Classification was done on breast cancers according to ER status (A:GSE20194 and B:GSE4922); on muscle disease samples according to gender (C) or to the muscle disease type (D), one class vs. the rest. The solid grey line uses the samples assigned the highest SNR (and so likely to be biological outliers) by SNAGEE.
lowest quality samples is shown. Quality of the samples was determined with SNAGEE (solid line), MDQC (dashed line), GNUSE (dotted line), NUSE (grey dotted line) and RLE (dash-dot line). Classification was done on breast cancers according to ER status (A:GSE20194 and B:GSE4922); on muscle disease samples according to gender (C) or to the muscle disease type (D), one class vs. the rest. The solid grey line uses the samples assigned the highest SNR (and so likely to be biological outliers) by SNAGEE.
References
-
- Wilson CL, Miller CJ (2005) Simpleaffy: a bioconductor package for affymetrix quality control and data analysis. Bioinformatics 21: 3683–3685. - PubMed
-
- Dunning MJ, Smith ML, Ritchie ME, Tavare S (2007) beadarray: R classes and methods for illumina bead-based data. Bioinformatics 23: 2183–2184. - PubMed
-
- Cohen Freue GV, Hollander Z, Shen E, Zamar RH, Balshaw R, et al. (2007) Mdqc: a new quality assessment method for microarrays based on quality control reports. Bioinformatics 23: 3162–3169. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

