Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure
- PMID: 23251478
- PMCID: PMC3521004
- DOI: 10.1371/journal.pone.0051274
Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure
Abstract
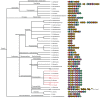
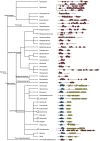
Escherichia coli and a few other members of the Enterobacteriales can produce functional amyloids known as curli. These extracellular fibrils are involved in biofilm formation and studies have shown that they may act as virulence factors during infections. It is not known whether curli fibrils are restricted to the Enterobacteriales or if they are phylogenetically widespread. The growing number of genome-sequenced bacteria spanning many phylogenetic groups allows a reliable bioinformatic investigation of the phylogenetic diversity of the curli system. Here we show that the curli system is phylogenetically much more widespread than initially assumed, spanning at least four phyla. Curli fibrils may consequently be encountered frequently in environmental as well as pathogenic biofilms, which was supported by identification of curli genes in public metagenomes from a diverse range of habitats. Identification and comparison of curli subunit (CsgA/B) homologs show that these proteins allow a high degree of freedom in their primary protein structure, although a modular structure of tightly spaced repeat regions containing conserved glutamine, asparagine and glycine residues has to be preserved. In addition, a high degree of variability within the operon structure of curli subunits between bacterial taxa suggests that the curli fibrils might have evolved to fulfill specific functions. Variations in the genetic organization of curli genes are also seen among different bacterial genera. This suggests that some genera may utilize alternative regulatory pathways for curli expression. Comparison of phylogenetic trees of Csg proteins and the 16S rRNA genes of the corresponding bacteria showed remarkably similar overall topography, suggesting that horizontal gene transfer is a minor player in the spreading of the curli system.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

