Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization
- PMID: 23251644
- PMCID: PMC3522617
- DOI: 10.1371/journal.pone.0051862
Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization
Abstract
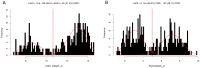
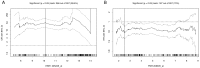
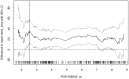
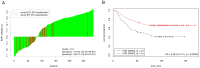
Gene or protein expression data are usually represented by metric or at least ordinal variables. In order to translate a continuous variable into a clinical decision, it is necessary to determine a cutoff point and to stratify patients into two groups each requiring a different kind of treatment. Currently, there is no standard method or standard software for biomarker cutoff determination. Therefore, we developed Cutoff Finder, a bundle of optimization and visualization methods for cutoff determination that is accessible online. While one of the methods for cutoff optimization is based solely on the distribution of the marker under investigation, other methods optimize the correlation of the dichotomization with respect to an outcome or survival variable. We illustrate the functionality of Cutoff Finder by the analysis of the gene expression of estrogen receptor (ER) and progesterone receptor (PgR) in breast cancer tissues. This distribution of these important markers is analyzed and correlated with immunohistologically determined ER status and distant metastasis free survival. Cutoff Finder is expected to fill a relevant gap in the available biometric software repertoire and will enable faster optimization of new diagnostic biomarkers. The tool can be accessed at http://molpath.charite.de/cutoff.
Conflict of interest statement
Figures

References
-
- Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, et al. (1993) Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst 85: 200–206. - PubMed
-
- Harvey JM, Clark GM, Osborne CK, Allred DC (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 17: 1474–1481. - PubMed
-
- Remmele W, Stegner HE (1987) Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (er-ica) in breast cancer tissue. Pathologe 8: 138–140. - PubMed
-
- von Minckwitz G, Müller BM, Loibl S, Budczies J, Hanusch C, et al. (2011) Cytoplasmic poly(adenosine diphosphate-ribose) polymerase expression is predictive and prognostic in patients with breast cancer treated with neoadjuvant chemotherapy. J Clin Oncol 29: 2150–2157. - PubMed
-
- Mazumdar M, Glassman JR (2000) Categorizing a prognostic variable: review of methods, code for easy implementation and applications to decision-making about cancer treatments. Stat Med 19: 113–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

