P2X receptors as drug targets
- PMID: 23253448
- PMCID: PMC3608433
- DOI: 10.1124/mol.112.083758
P2X receptors as drug targets
Abstract
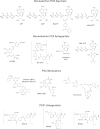
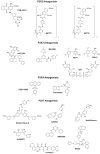
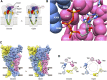
The study of P2X receptors has long been handicapped by a poverty of small-molecule tools that serve as selective agonists and antagonists. There has been progress, particularly in the past 10 years, as cell-based high-throughput screening methods were applied, together with large chemical libraries. This has delivered some drug-like molecules in several chemical classes that selectively target P2X1, P2X3, or P2X7 receptors. Some of these are, or have been, in clinical trials for rheumatoid arthritis, pain, and cough. Current preclinical research programs are studying P2X receptor involvement in pain, inflammation, osteoporosis, multiple sclerosis, spinal cord injury, and bladder dysfunction. The determination of the atomic structure of P2X receptors in closed and open (ATP-bound) states by X-ray crystallography is now allowing new approaches by molecular modeling. This is supported by a large body of previous work using mutagenesis and functional expression, and is now being supplemented by molecular dynamic simulations and in silico ligand docking. These approaches should lead to P2X receptors soon taking their place alongside other ion channel proteins as therapeutically important drug targets.
Figures

References
-
- Ahlquist RP. (1948) A study of the adrenotropic receptors. Am J Physiol 153:586–600 - PubMed
-
- Albers-Schönberg G, Arson BH, Chabala JC, Douglas AW, Eskola P, Fisher MH, Lusi A, Mrozik H, Smith JL, Tolman RL. (1981) Avermectins: structure determination. J Am Chem Soc 103:4216–4221
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

