HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency
- PMID: 23253934
- PMCID: PMC3558390
- DOI: 10.1186/1742-4690-9-111
HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency
Abstract
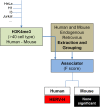
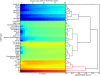
Background: Certain post-translational modifications to histones, including H3K4me3, as well as binding sites for the transcription factor STAT1, predict the site of integration of exogenous gamma-retroviruses with great accuracy and cell-type specificity. Statistical methods that were used to identify chromatin features that predict exogenous gamma-retrovirus integration site selection were exploited here to determine whether cell type-specific chromatin markers are enriched in the vicinity of endogenous retroviruses (ERVs).
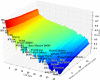
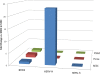
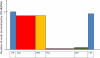
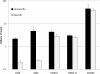
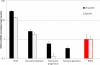
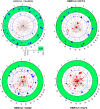
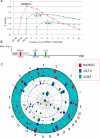
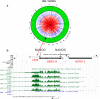
Results: Among retro-elements in the human genome, the gamma-retrovirus HERV-H was highly associated with H3K4me3, though this association was only observed in embryonic stem (ES) cells (p < 10-300) and, to a lesser extent, in induced pluripotent stem (iPS) cells. No significant association was observed in nearly 40 differentiated cell types, nor was any association observed with other retro-elements. Similar strong association was observed between HERV-H and the binding sites within ES cells for the pluripotency transcription factors NANOG, OCT4, and SOX2. NANOG binding sites were located within the HERV-H 5'LTR itself. OCT4 and SOX2 binding sites were within 1 kB and 2 kB of the 5'LTR, respectively. In keeping with these observations, HERV-H RNA constituted 2% of all poly A RNA in ES cells. As ES cells progressed down a differentiation pathway, the levels of HERV-H RNA decreased progressively. RNA-Seq datasets showed HERV-H transcripts to be over 5 kB in length and to have the structure 5'LTR-gag-pro-3'LTR, with no evidence of splicing and no intact open reading frames.
Conclusion: The developmental regulation of HERV-H expression, the association of HERV-H with binding sites for pluripotency transcription factors, and the extremely high levels of HERV-H RNA in human ES cells suggest that HERV-H contributes to pluripotency in human cells. Proximity of HERV-H to binding sites for pluripotency transcription factors within ES cells might be due to retention of the same chromatin features that determined the site of integration of the ancestral, exogenous, gamma-retrovirus that gave rise to HERV-H in the distant past. Retention of these markers, or, alternatively, recruitment of them to the site of the established provirus, may have acted post-integration to fix the provirus within the germ-line of the host species. Either way, HERV-H RNA provides a specific marker for pluripotency in human cells.
Figures


References
-
- Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10:395–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

